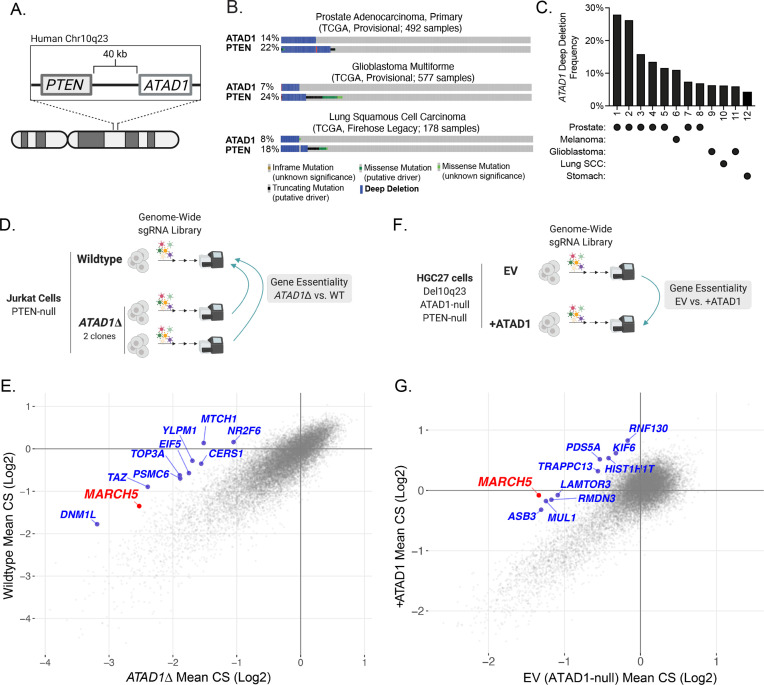
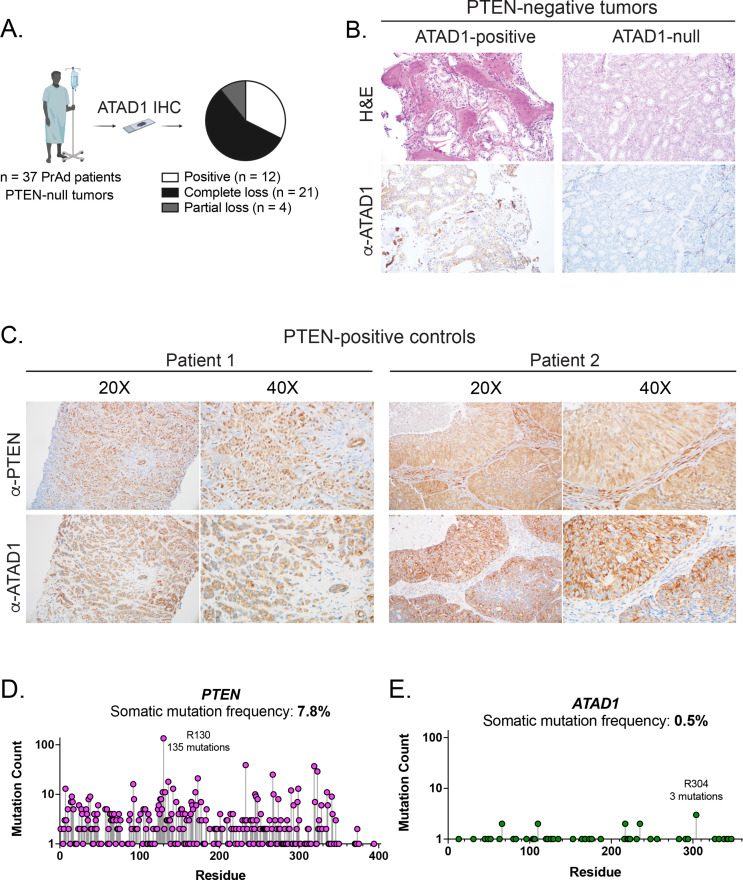
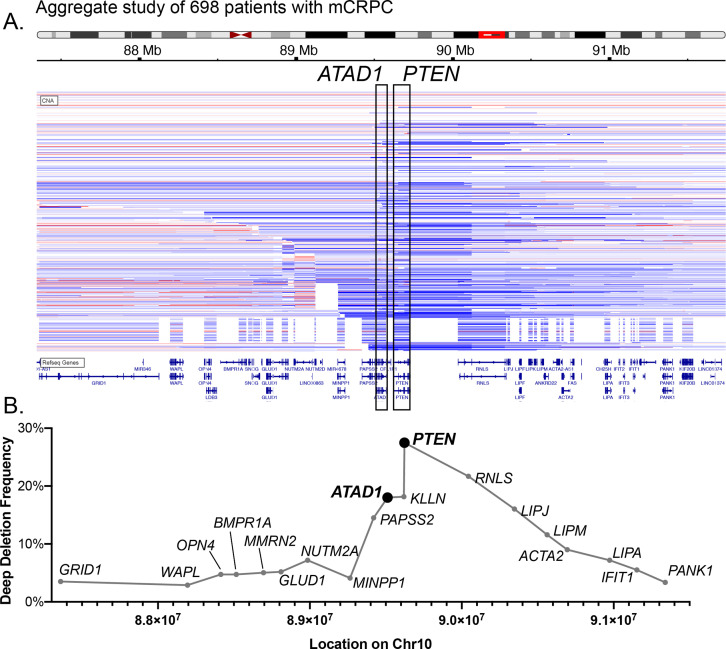
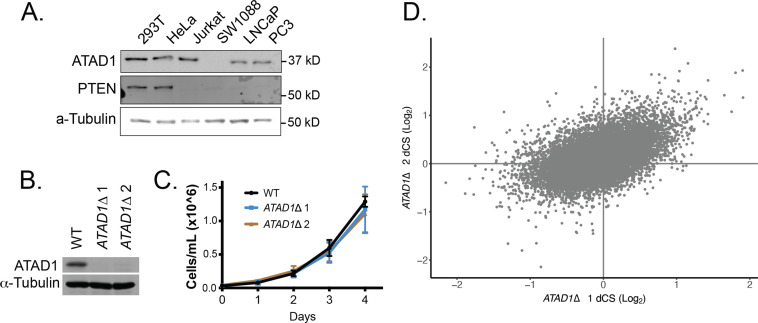
Figure 1. ATAD1 is co-deleted with PTEN in cancer and its loss confers synthetic lethal vulnerabilities.
(A) Schematic of PTEN and ATAD1 loci. (B) Oncoprint plots from three TCGA studies of cancer. ATAD1 and PTEN alteration frequencies are shown, with blue bars indicating deep deletions. (C) Frequency of ATAD1 deep deletions across various cancer types; data from cBioPortal. (D) CRISPR screen design for wild-type (WT) and ATAD1∆ Jurkat cells. (E) Jurkat CRISPR screen results; each point represents one gene. CRISPR score (CS) values were calculated by taking the average log2 fold-change in relative abundance of all sgRNAs targeting a given gene over 14 population doublings. WT CS values are shown on the y-axis. The CS values per gene for each of the two ATAD1∆ clones were averaged and are plotted on the x-axis. The top 10 genes that were differentially essential between WT and ATAD1∆ are labeled in blue, with MARCH5 labeled in red. (F) CRISPR screen design for HGC27 cells (Chr10q23 deletion, ATAD1-null) comparing gene essentiality in ATAD1 complemented cells or empty vector (EV) (ATAD1-null) control. (G) HGC27 CRISPR screen results; CS values are as described for (E). The x-axis depicts CS for the ATAD1-null condition of EV-transduced cells, and the y-axis depicts CS for the ATAD1-complemented (+ATAD1) condition. Labels are as described for (E).