Figure 3. mAb 769C2 defines a site of vulnerability with antibody contacts on both gH and gL molecules.
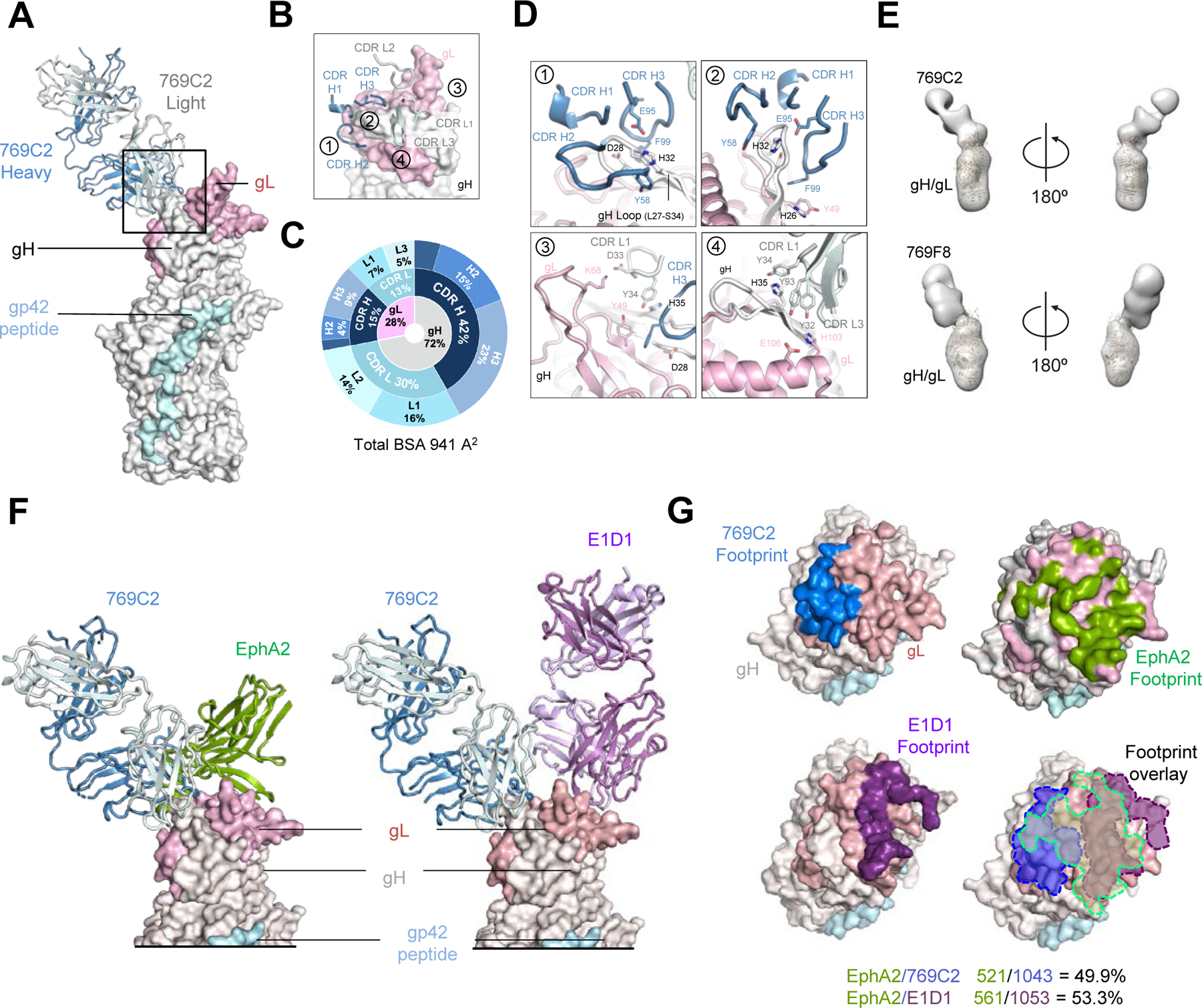
(A) Structure of 769C2 Fab (ribbon representation, heavy chain and light chain are colored with sky blue and light blue respectively) in complex with EBV gH/gL/gp42peptide (surface representation; gH, gL, and gp42p are colored in light grey, light pink, and pale cyan respectively) are shown.
(B) Major antibody loops involved in gH/gL recognition are shown in ribbon representation with gH/gL in surface representation.
(C) CDR-glycoprotein epitope BSA % are shown represented in pie-chart format. The 769C2 epitope is made up of 72% gH and 28% gL (inner circle), with both heavy and light chain recognition of gH and gL (middle), with multiple CDR loops used to recognize the gH/gL molecule (outer).
(D) mAb 769C2 contact residues are shown in stick representations based on (1, 2) CDR H1, H2 and H3; (3) CDR L1 and H3; (4) CDR L1 and L3.
(E) Negative-stain EM maps of 769C2 (top) and competing mAb 769F8 (bottom) in complex with EBV gH/gL. The EBV gH/gL/gp42p structure is fitted into the electron density for reference.
(F) Structural comparison of gH/gL/gp42p (surface representation) in complex with 769C2 or EphA2 (left) or E1D1 (right) respectively.
(G) Comparison of the 769C2 epitope (blue, upper left) to mAb E1D1 epitope (purple, lower left) or the EphA2 (bright green, top right) binding site with all shown overlaid (lower right). The % BSA that is identical between the EphA2 binding site (top) and the antibody epitopes is indicated at the base of the figure.
