Figure 5. Antigenic sites of vulnerability on the EBV gH/gL and gH/gL/gp42 fusion machinery.
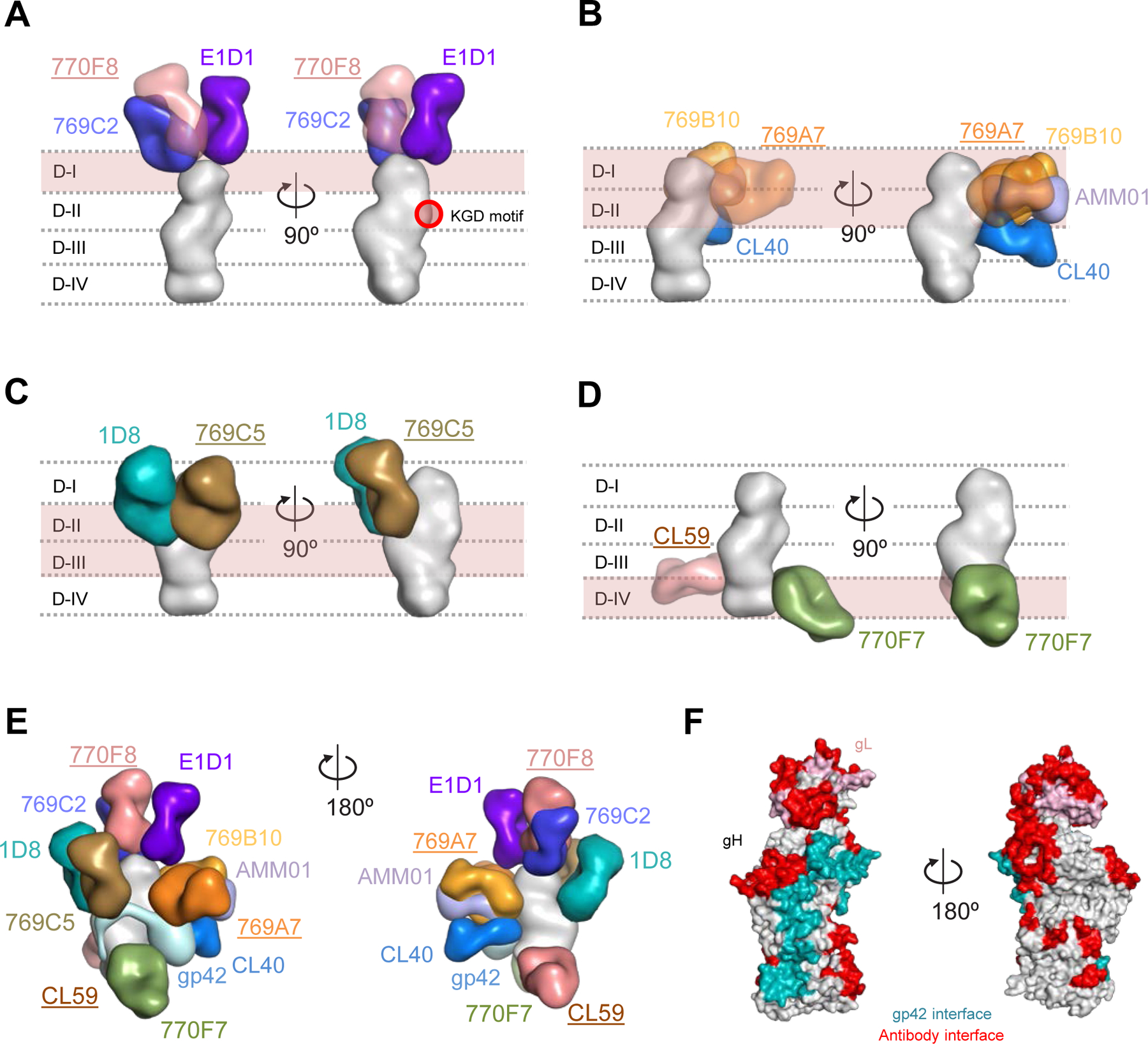
Structure-based composite model of EBV gH/gL and gH/gL/gp42 with neutralizing antibodies. EBV gH domains are indicated by dotted lines for (A-D). Antibody names that are underlined indicate that the structural information is provided by negative-stain EM maps and antibody docking. Antibodies and gH/gL molecules are shown in surface representation.
(A) mAbs 769C2 (blue), 770F8 (rose), and E1D1 (purple) targeting the gH/gL D-I region.
(B) mAbs 769B10 (orange), 769A7 (gold), AMMO1 (periwinkle blue), and CL40 (blue) targeting gH/gL D-I and D-II.
(C) mAbs 1D8 (teal), and 769C5 (brown) targeting the gH/gL D-II and D-III region.
(D) mAbs CL59 (salmon), and 770F7 (green), targeting the gH/gL D-IV region.
(E) EBV glycoproteins gH/gL/gp42 (surface representation) are shown bound to eleven mAbs. The antibody-antigen model is shown in two orientations to allow visibility of the antibodies.
(F) Antibody and gp42 interfaces are shown on the gH/gL/gp42p molecules (surface representation).
See also Figures S4-S5.
