Figure 6. Comparison of herpesvirus antigenic sites of vulnerability on the gH/gL fusion machinery.
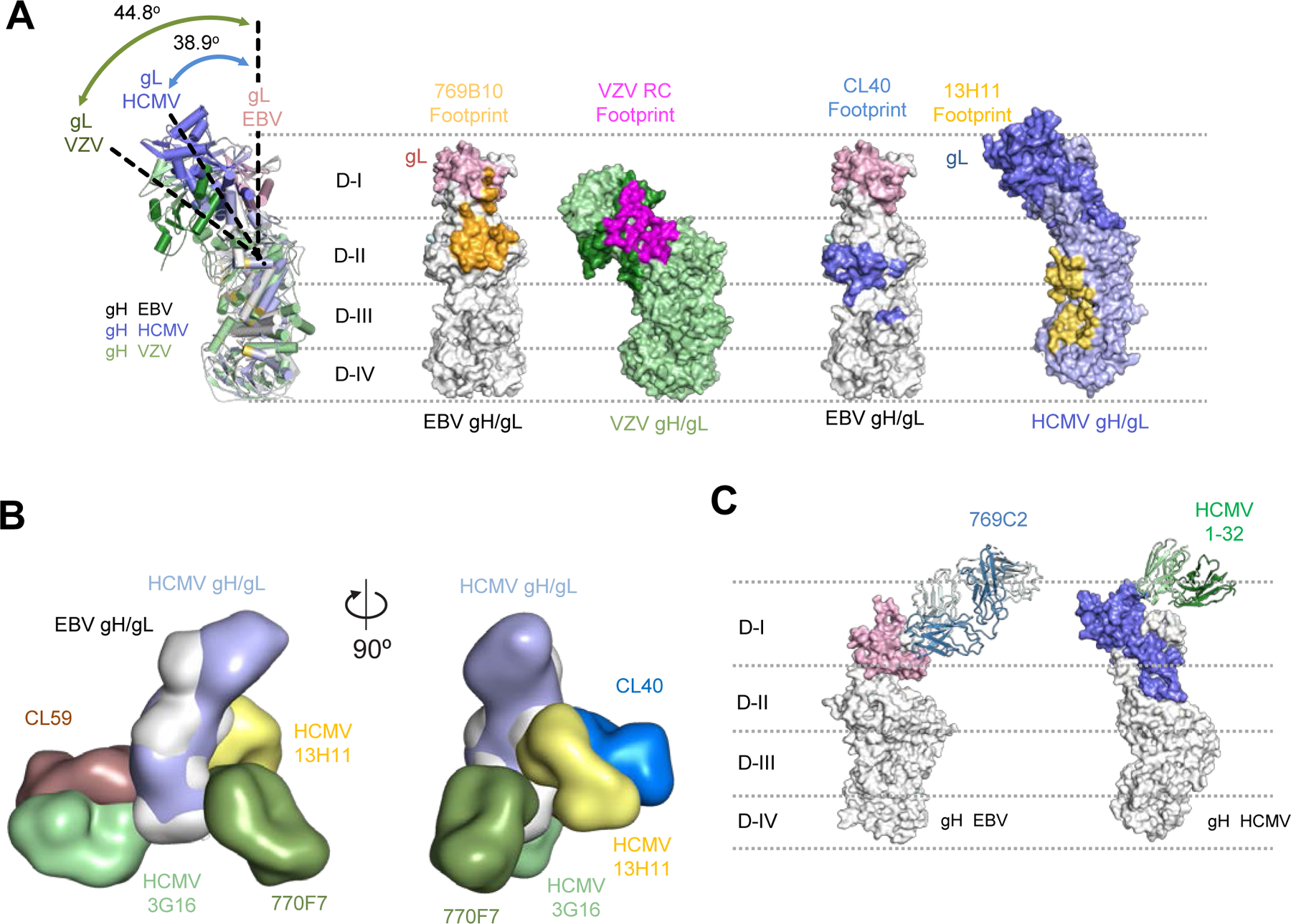
(A) Comparisons of the 796B10 epitope to VZV RC (purple), EBV CL40 (blue), and HCMV 13H11 (yellow) antibody epitopes are mapped on the respective viral gH/gL molecules, shown in surface representation. The EBV gH/gL structural overlap (left) with VZV and HCMV gH/gL molecules indicates the different orientation or tilt of the gH D-I/gL domains in each molecule relative to each other. The approximate location of the gH DI-DIV domains are indicated by horizontal dotted lines.
(B) Overlay of HCMV and EBV gH/gL molecules with HCMV mAbs 3G16, 13H11 and EBV mAbs 770F7, CL40, and CL59 shown in surface representation with two orientations.
(C) Comparison of mAb 796C2-gH/gL recognition to HCMV gH/gL-targeting mAb 1–32 (green) shown in ribbon representation, with herpesvirus gH/gL molecules shown in surface representation. The approximate location of the gH DI-DIV domains are indicated by horizontal dotted lines.
See also Figure S5.
