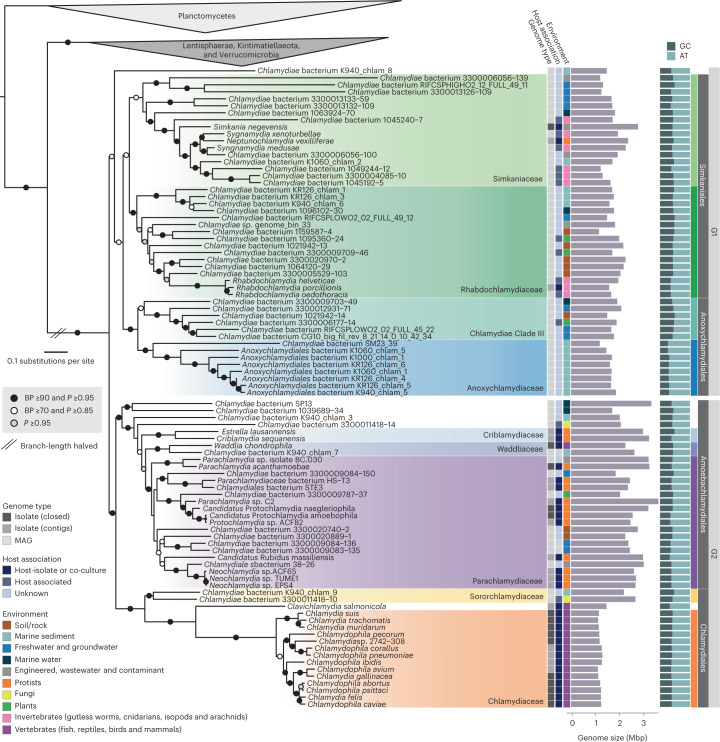
Fig. 1. Robust species phylogeny of PVC bacteria.
Concatenated (74 marker genes) Bayesian phylogeny of 180 PVC bacteria with compositionally heterogeneous sites removed (8,151 amino acid sites). Circles indicate bipartition support from posterior probability (P) (CAT + GTR + Γ4 model), and nonparametric bootstraps (BP) (LG + C60 + F + Γ4-derived PMSF approximation). The tree is rooted by Planctomycetes. Reduced branch lengths are indicated by parallel lines, and substitutions per site by the scale bar. Genome type, evidence for host association and the environment from which the genome was obtained are indicated by coloured squares according to the legend. Genome size (Mbp in purple) and GC content (%GC in dark blue and %AT in light blue) are indicated by bars. Higher-level taxonomic classifications are indicated and chlamydial families outlined by coloured boxes. See also Supplementary Data 2 for genome characteristics and Supplementary Data 6 for the uncollapsed species phylogeny.