FIGURE 2.
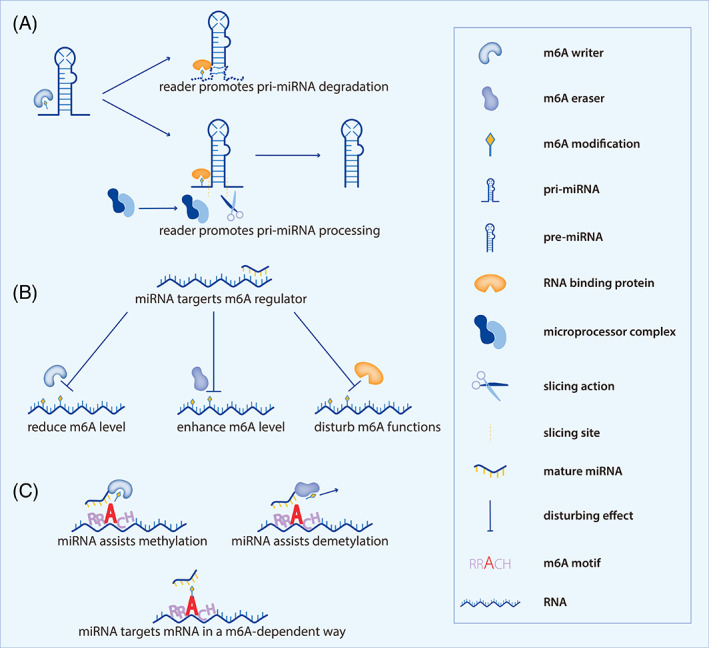
The patterns of interaction between m6A modification and miRNAs. (A) m6A modification mediates pri‐miRNAs processing. m6A writer deposits methyl at the m6A motif of pri‐miRNA followed by m6A reader recognizing it. Part of readers like HNRNP2B1, NKAP and YTHDC1 recruit microprocessor complex for flanking slicing, subsequently receiving a precursor miRNA, while YTHDF2 could accelerate pre‐miRNA degradation. (B) miRNAs target 3′s UTR of m6A regulators. miRNAs could induce m6A level alteration or impede m6A reader function implementation through targeting m6A regulators' 3′ UTR. (C) miRNAs orchestrate with m6A regulators to exert their intrinsic functions. miRNAs could assist m6A methylation and demethylation by interacting with the writer and eraser respectively. miRNA targets mRNA 3′UTR in an m6A‐dependent way
