Figure 4. Transcriptional responses of enterocytes correlate with improved lipid absorption and transport.
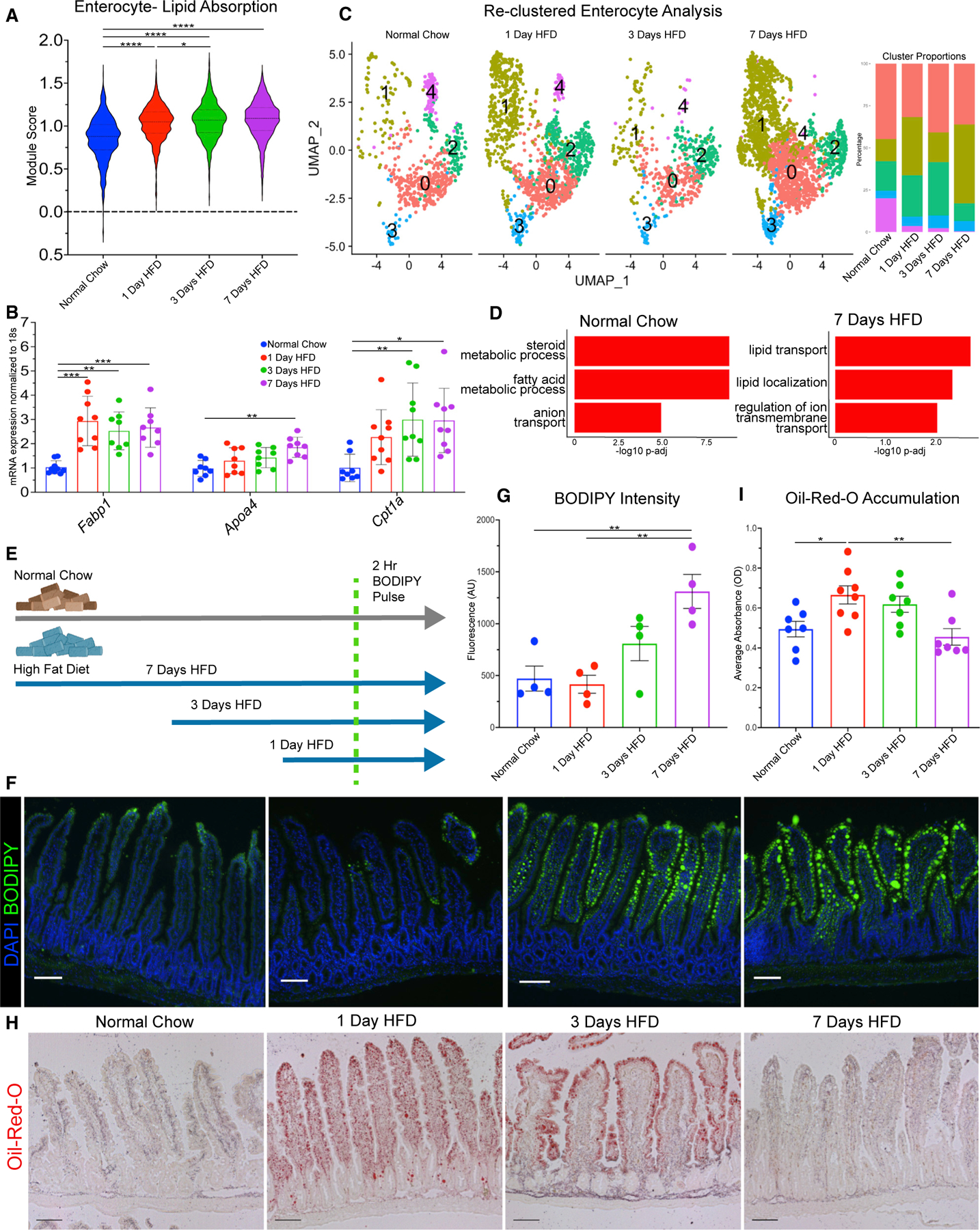
(A) Lipid absorption score compared between enterocytes in each dietary condition.
(B) qRT-PCR between dietary conditions for fatty acid binding protein 1 (Fabp1), apolipoprotein A-IV (Apoa4), and carnitine palmitoyltransferase 1a (Cpt1a).
(C) Uniform manifold approximation and projection (UMAP) of enterocytes reclustered into 5 subsets: 0 (1,735 cells), 1 (1,671 cells), 2 (847 cells), 3 (281 cells), and 4 (200 cells). Barplot visualizes cluster proportion colored by cluster number. Breakdown per condition: normal chow (675 cells), 1 day of HFD (1,308 cells), 3 days of HFD (549 cells), and 7 days of HFD (2,202 cells).
(D) Top 3 upregulated Gene Ontology terms (Biological Process) for normal chow and 7 days of HFD in enterocyte subclusters −0 and −1.
(E) Diagram of BODIPY pulse-chase experiment. Normal chow, gray arrow; HFD, blue arrow; green dashed line denotes time when all mice were gavaged with BODIPY and analyzed 2 h later.
(F) Fluorescence images of BODIPY (green) lipid absorbance in proximal jejunum 2 h post-gavage. DAPI counterstains nuclei in blue. Scale bars: 100 μm.
(G) Quantification of BODIPY fluorescence intensity. n = 4 per condition.
(H) Oil red O staining of proximal jejunum for each condition. Scale bars: 100 μm.
(I) Quantification of (H) reported as optical density (OD) for oil red O accumulation. n = 7 for each condition. (A, F, and H) Error bars are SEM. One-way ANOVA; ns, not significant, *p < 0.05, **p < 0.005, ****p < 0.0001.
