Abstract
The utility of using severe-acute respiratory syndrome coronavirus-2 (SARS-CoV-2) RNA for assessing the prevalence of COVID-19 within communities begins with the design of the sample collection program. The objective of this study was to assess the utility of 24-hour composites as representative samples for measuring multiple microbiological targets in wastewater, and whether normalization of SARS-CoV-2 by endogenous targets can be used to decrease hour to hour variability at different watershed scales. Two sets of experiments were conducted, in tandem with the same wastewater, with samples collected at the building, cluster, and community sewershed scales. The first set of experiments focused on evaluating degradation of microbiological targets: SARS-CoV-2, Simian Immunodeficiency Virus (SIV) – a surrogate spiked into the wastewater, plus human waste indicators of Pepper Mild Mottle Virus (PMMoV), Beta-2 microglobulin (B2M), and fecal coliform bacteria (FC). The second focused on the variability of these targets from samples, collected each hour on the hour. Results show that SARS-CoV-2, PMMoV, and B2M were relatively stable, with minimal degradation over 24-h. SIV, which was spiked-in prior to analysis, degraded significantly and FC increased significantly over the course of 24 h, emphasizing the possibility for decay and growth within wastewater. Hour-to-hour variability of the source wastewater was large between each hour of sampling relative to the variability of the SARS-CoV-2 levels calculated between sewershed scales; thus, differences in SARS-CoV-2 hourly variability were not statistically significant between sewershed scales. Results further provided that the quantified representativeness of 24-h composite samples (i.e., statistical equivalency compared against hourly collected grabs) was dependent upon the molecular target measured. Overall, improvements made by normalization were minimal within this study. Degradation and multiplication for other targets should be evaluated when deciding upon whether to collect composite or grab samples in future studies.
Keywords: SARS-CoV-2, Grab, Composite, Degradation, Hour-to-hour variability, Normalization
Graphical abstract
1. Introduction
Wastewater-based epidemiology (WBE) has become a global standard for the effective tracking of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), the virus responsible for the COVID-19 pandemic impacting communities world-wide (Betancourt et al., 2021; Haramoto et al., 2020; Randazzo et al., 2020; Rosiles-Gonzalez et al., 2021; Schmitz et al., 2021; Sherchan et al., 2020; Westhaus et al., 2021; Zhan et al., 2022; Wilder et al., 2021). Surveillance and monitoring programs have been increasingly established, internationally, to illustrate spatial and temporal trends of disease presence from contributing individuals within communities (Betancourt et al., 2021; Randazzo et al., 2020; Rosiles-Gonzalez et al., 2021; Schmitz et al., 2021; Sherchan et al., 2020; Westhaus et al., 2021; Gerrity et al., 2021; Hart and Halden, 2020; Vo et al., 2022). It has been shown that there is a predictive correlation (2 to 10 days) between viral loads found in wastewater with community prevalence of illness and with incidences of hospital cases (Solo-Gabriele et al., 2022; Wu et al., 2022). This has allowed for further investigations to define the usefulness of WBE to track the spread of SARS-CoV-2 within a community and inform policy decisions for managing the status of SARS-CoV-2 for the corresponding sewershed. However, many studies have found limitations in the use of broad-based WBE for detection and accurate reporting of SARS-CoV-2 due to a variety of factors, inclusive of identifying appropriate sampling strategies (Ahmed et al., 2020a; Ahmed et al., 2021; Ahmed et al., 2022; Bivins et al., 2020a; Kantor et al., 2021; Bertels et al., 2022).
As described by others previously (George et al., 2022; Grijalva et al., 2022), within WBE there are two primary sampling strategies for the appropriate collection of wastewater, grab sampling and composite sampling. Depending upon the study, both strategies are utilized for the effective collection of wastewaters and may provide different results. The variability seen with grab sampling is typically higher than that of composite sampling in that the composition of wastewater in a sewer system, consisting of input waste from various sources can depend upon human behavior, and downstream results can correspond to when a specific sample is collected. Composite samples, conversely, are considered ‘averaged’ in that they are comprised of aliquots, collected over a specific time frequency, to adjust for the variability in wastewater composition. For composite sample types, degradation is predicted to be a compounding factor in that the wastewater sample remains in the sample bottle for a period of time prior to analysis and its properties can change during this time. For example, for a 24-h composite sample collected hourly, the first aliquot is held for 24 h prior to analysis during which the microbiological signal can potentially degrade.
The degradation of SARS-CoV-2 within the sewer system has been previously linked to water quality parameters – such as dissolved oxygen, turbidity, or pH (Bertels et al., 2022; Li et al., 2021), as well as, specifically, the temperature of wastewater (Hart and Halden, 2020; Ahmed et al., 2020b; Bivins et al., 2020b; McCall et al., 2022; Schussman and McLellan, 2022; Weidhaas et al., 2021; Yang et al., 2022). McCall et al. (2022) through sewer transport modeling evaluated the impact that microbial degradation plays within WBE. McCall et al. (2022) compared degradation rates as a function of temperature and found significant ranges for studies evaluating decay at two similar temperatures: 0.18/h at 35 °C (Weidhaas et al., 2021) and 0.012/h at 37 °C (Ahmed et al., 2020b). Possible differences in these decay rates were attributed to differences in wastewater composition, initial SARS-CoV-2 levels (Bivins et al., 2020b), and to differences in sample preparation. Of note, the study that reported 0.18/h degradation rate used endogenous SARS-CoV-2 for evaluation whereas the study that reported 0.012/h degradation used an exogenous SARS-CoV-2 spike. In these prior studies, the degradation rates of endogenous SARS-CoV-2 were higher than those of spiked samples, demonstrating slower decay of the exogenous virus. In the current study, the SARS-CoV-2 evaluated was endogenous. To further assess the possible influence of spiking viruses into wastewater, we added an external spike, Simian Immunodeficiency Virus (SIV), due to its availability from an internal laboratory. SIV is an enveloped RNA virus sharing some similar properties to SARS-CoV-2 and given that it is simian specific, would not be expected within domestic wastewater. To our knowledge, this is the first study which utilized SIV as a viral comparison for SARS-CoV-2 degradation within wastewater.
In addition to water quality and sample preparation, the input population of individuals contributing to a sewershed is also an important parameter to consider when designing a sample collection program (Bertels et al., 2022; Grijalva et al., 2022). A review by Bertels et al. (2022) describes findings from Wu et al. (2022) and Wilder et al. (2021) in which the correlations between SARS-CoV-2 in wastewater and COVID-10 cases strengthens as population or sewershed size increases. Larger contributing populations can result in less variability in the wastewater composition due to the averaging effects of people's activities over larger numbers; however assuming a static population count remains a drawback to WBE wherein human activity plays an important role in the viral load being deposited within the sewer system on a given day (Bertels et al., 2022). Counter arguments can include that large community-wide WWTPs can consist of more ‘types’ of water than smaller-scale sewer systems from residential, commercial, and industrial activities, thereby contributing towards variability and either over- or under-reporting of SARS-CoV-2 incidence against clinical cases. In addition, the distance that wastewater must travel from a drain to the downstream point of collection increases for larger service populations and it is believed that wastewater travel time can obscure the variability results and degradation occurring within wastewater samples for a specified target (McCall et al., 2022; Schussman and McLellan, 2022).
Intra-day variability of SARS-CoV-2 abundance has been predicted within the literature to fluctuate due to 1) the wastewater source and sampling frequency (Ahmed et al., 2021; Li et al., 2021), 2) the input population of the community shedding into a sewer system (Kantor et al., 2021; Grijalva et al., 2022; McCall et al., 2022; Bertanza et al., 2022; Song et al., 2021), 3) wide-variation of standardization approaches for quantification of viral particles (Ahmed et al., 2022; Kantor et al., 2021), and 4) watershed scale (George et al., 2022). For example, George et al. (2022) found that the smaller the watershed scale the larger the variability of the SARS-CoV-2 signal on an hour-to-hour basis. As a result, George et al. (2022) recommends higher frequency sampling for smaller sewersheds in order to obtain a representative sample. As wastewater is a combination of various water sources which drain from a building, not all water (and corresponding solids which are typically linked to more SARS-CoV-2) draining into the sewer system is expected to contain SARS-CoV-2 (Kim et al., 2022). Sinks, toilets, showers, dishes and clothing washing as well as other sources of drainage are all ‘input sources’ for pathogens to enter the sewer system (Gibas et al., 2021). Some water sources are expected to contain higher levels of SARS-CoV-2, such as those containing nasal discharge and sputum from face washing, and feces as well as urine from individuals carrying the virus. To accommodate for these differences in water types, some studies have normalized the SARS-CoV-2 signal (Zhan et al., 2022; Wilder et al., 2021; Ahmed et al., 2022). The normalized signal is the ratio of SARS-CoV-2 concentrations divided by the concentration of the human waste indicator. In this study, the commonly used indicators of human waste evaluated included the dietary-originating plant pathogen, Pepper Mild Mottle Virus (PMMoV) (Ahmed et al., 2021; Kantor et al., 2021; Symonds et al., 2016), and fecal coliform bacteria (FC) (Gerrity et al., 2021; Symonds et al., 2016; Davoodi et al., 2018; Zhang et al., 2021). In addition, this study also evaluated a less common indicator, the Beta-2 microglobulin (B2M) gene mRNA found in human cells (Zhan et al., 2022; Gussow et al., 1987; Sharkey et al., 2021), for normalization of the SARS-CoV-2 signal.
Overall, the goal of this study was to add to the existing literature by simultaneously evaluating the influence of sample hold times (degradation) and wastewater variability on the ability of composite samples to represent a daily average. Our approach was to build upon the work of George et al. (2022) and Grijalva et al. (2022) by conducting both degradation experiments coupled with an assessment of the hour-to-hour variability by sewershed scale by collecting grab samples and creating a composite for later molecular comparison. Our study is unique in that it simultaneously evaluated hour-to-hour variability of the sewage by grab sampling, along with its degradation within laboratory-created composite samples. Using this simultaneous approach, we were able to assess the combined effects of degradation and variability of the wastewater source using the same original wastewater sample. Only through this approach were we able to evaluate the impact of the hourly hold times on the microbe levels within the composite samples using the same wastewater therefore controlling for potential differences in wastewater quality. We augmented this assessment by including measurements of water quality, human waste indicators, and inclusion of an exogenous virus. The study is also unique in the inclusion of multiple microbial measures which provided insights on potential variabilities between microbial targets and allowed for the assessment of SARS-CoV-2 normalization using three human waste indicators. The physical chemical properties of the wastewater source were also documented to provide details necessary for comparison with other studies, given that the stability of microbial targets has been documented in prior studies to also be influenced by water quality.
2. Materials and methods
2.1. Sample collection
Three pairs of experiments for a total of six experiments were conducted as part of this study. Each pair corresponded to a different sewershed scale. The first pair (experiment 1A and 1B, 9:00 am on April 22nd until 9:00 am on April 23rd, 2021) corresponded to the building scale with samples collected from a wastewater sewer manhole at the University of Miami Gables Campus that serviced a dormitory that housed about 500 students. The second pair (experiment 2A and 2B, 7:00 am on April 8th until 7:00 am on April 9th, 2021) corresponded to the cluster scale, also at the University of Miami Gables campus, but the samples were collected from a lift station that serviced four residential buildings housing 1400 students plus 20 office and academic buildings. The third pair (experiment 3A and 3B, 7:00 am on June 3rd until 8:00 am on June 4th, 2021) was collected from the raw sewage line upstream of the grit chamber at the Miami-Dade County Central District Wastewater Treatment Plant (CDWWTP); the CDWWTP services a population of roughly 830,000 individuals. Upon sample collection, date and time were recorded as well as general ambient conditions (air temperature and humidity) as reported by the NavClock iPhone app (Table 1 ).
Table 1.
Summarized water quality parameters per experiment and sewershed scale for all collected wastewater samples. Each value is an average of n = 24 grab samples, recorded each hour on the hour for both Experiments “A” and “B”.
| Water Quality Parameter (n = 24 grab average) | Experiments 1-3A |
Experiments 1-3B |
||||
|---|---|---|---|---|---|---|
| Building | Cluster | Community | Building | Cluster | Community | |
| pH Field | 8.68 | 7.83 | 6.98 | 8.20 | 8.24 | 7.23 |
| pH Lab | 8.71 | 7.77 | 7.10 | 8.11 | 8.19 | 6.91 |
| Water Temperature (°C) | 22.1 | 22.0 | 22.4 | 24.1 | 24.2 | 25.0 |
| Specific Conductivity (μS/cm) | 4665 | 3086 | 14,940 | 3191 | 2586 | 14,560 |
| Turbidity (FNU) | 46.2 | 36.8 | 105.8 | 89.3 | 62.4 | 83.7 |
| Dissolved Oxygen (mg/L) | 2.2 | 2.1 | 2.0 | 6.0 | 4.7 | 2.4 |
| Air Temperature (°C) | N/Aa | N/A | 26.0 | 25.6 | 23.2 | 26.2 |
| Humidity (%) | N/A | N/A | 87.2 | 64.8 | 61.9 | 84.1 |
| Flow (m3/day) | N/A | N/A | N/A | 98.2 | 279 | 254,870 |
N/A = Not applicable.
The “A” set of experiments were designed to assess the degradation of the biological target. This set of samples, consisting of one large 16-l grab, was collected at the very beginning of each experiment. This 16-l grab was immediately split into 24 aliquots of subsamples for the analysis of microbial targets and water quality over time. The “B” set of experiments were designed to assess the hour-to-hour variability of wastewater at the point of sampling. These samples were collected manually, each hour over a 24-h timeframe per experiment, in 1-l bottles, then processed immediately for microbial targets, water quality, and primary concentration with minimal holding times from the time of collection (<30 min). Samples for experiments 1A/B and 2A/B were collected using a peristaltic pump and samples for experiment 3A/B were collected using a scooper attached to a long rod. Throughout sample collection, the sampling team adhered to the University's Environmental Health and Safety policies which required the use of personal protective equipment and disinfection of all items that came into contact with wastewater.
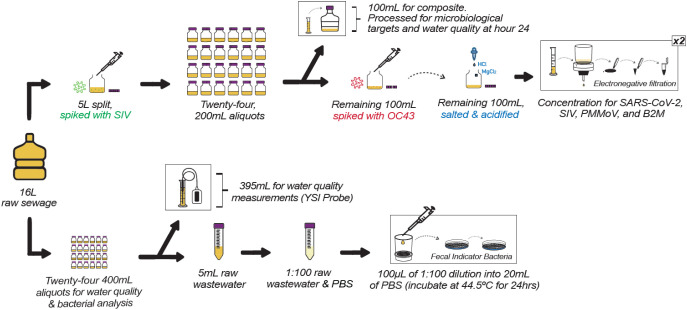
2.2. Sample splitting and initial spiking
All samples during splitting and composite preparation were kept at room temperature (22 °C). For the “A” set of experiments, 5 L of wastewater were removed from the 16-l grab sample collected at the start of each paired experiment (Fig. 1 ). To the 5 L subsample, a 500 μL spike of SIV (2.51 × 105 viral particles per μL), provided from an internal virology laboratory within the Center for AIDS Research at the University of Miami (UM), was added as a comparator; The 5 L volume was shaken vigorously and split into twenty-four 200 mL aliquots earmarked for concentration and molecular target analyses (SARS-CoV-2, SIV, PMMoV, and B2M) by Volcano 2nd Generation (V2G)-qPCR of RNA extracts. The remaining 11 L subsample was also immediately split upon collection into twenty-four 400 mL aliquots for water quality analyses and into twenty-four 5 mL aliquots placed into 15 mL sterile centrifuge tubes for FC analysis by culture.
Fig. 1.
Experimental “A” field and laboratory workflow visualized including sample splits, wastewater pre-treatment, and laboratory processing per aliquot of wastewater.
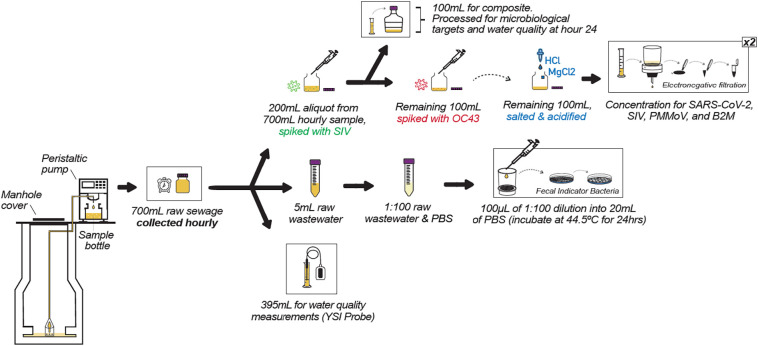
Simultaneously for the “B” set of experiments, the 1-l bottle (containing 700 mL of sample) brought back to the laboratory each hour was also split in a similar fashion as for experiment “A” (Fig. 2 ). Here the 1-l bottle was first shaken, then split into a 200 mL aliquot for concentration and microbial target analysis using molecular methods, a 5 mL sample placed in a 15 mL sterile centrifuge tube for fecal coliform analysis by culture, and 395 mL for water quality analysis. For the “B” set experiments, 20 μL of SIV spike were added to each of the 200 mL aliquots and shaken to homogenize.
Fig. 2.
Experimental “B” field and laboratory workflow visualized including sample splits, wastewater pre-treatment, and laboratory processing per aliquot of wastewater.
For each of the six experiments, composite samples were prepared manually in the laboratory from the 24-hourly samples. For each of the paired experiments, one container was used for the “A” set of samples and another container was used for the “B” set of samples. These composites were prepared from the 200 mL splits containing the SIV spikes. The 200 mL samples were shaken vigorously, and 100 mL was poured into a 3-l container. At the end of the 24-h experiment the 3-l container was shaken vigorously, and splits were prepared including a 100 mL aliquot earmarked for analysis of microbial targets using molecular methods, 5 mL for FC by culture, and 400 mL for water quality analysis. This stand-alone composite sample, created in-house, was analyzed at the end of all six experiments as an additional sample, per experiment for grab vs. composite assessment. For experiment 3B, a second, additional composite sample was provided by the CDWWTP from their refrigerated autosampler (HACH AS950 fitted with an IO9000 for flow proportional sampling) which collected samples from midnight to midnight the prior day. Aliquots were also prepared from this second composite sample for the analysis of microbial targets (by molecular and culture methods) plus water quality using the same aliquot volumes as described above.
2.3. Sample analysis for non-molecular targets
The 400 mL sample splits (each hourly sample plus composites) earmarked for water quality analyses were analyzed for water temperature (°C), specific conductivity (μS/cm), salinity (ppt), pH, dissolved oxygen (mg/L), and turbidity (FNU) using a pre-calibrated sonde (Xylem/YSI ProDSS). Averages for each biological parameter, per sewershed, are available within Table 1 and supporting information discussing water quality results are within the Supplemental Text. The 5 mL sample splits for FC analyses were analyzed by membrane filtration method using mFC agar (Hachich et al., 2012; Mahmud et al., 2019). Wastewater samples were diluted 100:1 in sterile phosphate buffered saline and 0.1 mL aliquots were then added to 20 mL sterile phosphate buffered saline, filtered, and incubated as per standard methods (Method 9222D, APHA et al., 2005). Colonies with characteristic blue color were counted as colony forming units (CFU).
2.4. Wastewater sample concentration for molecular analysis
The remaining 100 mL from the 200 mL subsamples (from each of the hourly and composite samples) were concentrated in real-time of collection, each hour, using electronegative filtration (EN) (Gibas et al., 2021; Sharkey et al., 2021; Babler et al., 2022; Juel et al., 2021). The process involved adding a process recovery control (20 μL of a heat inactivated (15 min @ 56 °C) human coronavirus-OC43 (OC43) at 10,090 particles/μL) (Kantor et al., 2021; Peccia et al., 2020; Zulli et al., 2021), followed by the addition of 51 % (w/v) MgCl2 (RICCA Chemical Company) to a concentration of 50 mM. OC43 has been used in WBE studies of COVID-19 as an effective process recovery control (Kantor et al., 2021; Sharkey et al., 2021; Babler et al., 2022; Peccia et al., 2020; Zulli et al., 2021; Pecson et al., 2021; Philo et al., 2021; Philo et al., 2022). As part of the EN process, samples were acidified with 10 % hydrochloric acid (Spectrum Chemical MFG Corp) until the pH dropped to between 3.5 and 4.5, as per standard methods (Method 9510, APHA 2017) (APHA et al., 2017). Wastewater in volumes of 30 to 50 mL was filtered through two separately processed electronegative membranes (47 mm diameter, 0.45 μm pore size, Millipore HAWP4700) per sample (Zhan et al., 2022; Sharkey et al., 2021; Babler et al., 2022; Abdelzaher et al., 2009; Abdelzaher et al., 2008). Concentrates were then prepared for each sample by folding filter membranes in on themselves four times and placing both filters immediately into 3 mL of 1× DNA/RNA Shield (Zymo) and storing at 4 °C, until subsequent RNA extraction (Zhan et al., 2022; Sharkey et al., 2021; Babler et al., 2022). All laboratory procedures handling wastewater and viral spikes were performed within a BioSafety Level 2 (BSL-2) laminar flow hood.
Concentrate samples were then split into 1 mL aliquots with one aliquot sent to the University of Miami Center for AIDS Research (CFAR) for molecular analysis with V2G-qPCR. The remaining 1 mL aliquots were stored at the University of Miami in −80 °C.
2.5. Viral RNA extraction
At the CFAR laboratory, RNA was extracted from 250 μL of the filter concentrate using a Quick-RNA™ Viral Kit (Zymo Research) and corresponding protocol, slightly modified in-house for the reduction of PCR inhibitors. In brief, wash buffer volumes and 100 % ethanol were increased from the 500 μL recommended volume to 650 μL for improved washing capability, and for the final elution, 10 μL of nuclease-free water, rather than the recommended 15 μL, was utilized to elute from the IC spin columns to reduce the amount of inhibitors passing through the columns alongside RNA. Following elution, a spike-in of Human Immunodeficiency Virus (HIV) RNA [~800 particles/μL] was added to each sample eluate, and a blank was created per sample set (10 μL nuclease-free water +30 μL HIV RNA) for proper comparative assessment. All RNA samples (n = 151) were stored at −20 °C following extraction (<1 week, to reduce drastic freeze-thaw) before analysis by V2G-qPCR. Following qPCR, RNA was stored at −80 °C.
2.6. V2G-qPCR analyses
Samples were analyzed for molecular targets via V2G-qPCR on a Bio-Rad CFX Connect Real-Time System (Bio-Rad Laboratories Inc., USA). The V2G assay, described in previous work (Zhan et al., 2022; Solo-Gabriele et al., 2022; Sharkey et al., 2021; Babler et al., 2022), was validated for detecting SARS-CoV-2 RNA early in the COVID-19 pandemic in saliva and has proven useful in detecting SARS-CoV-2 RNA, and other targets, from wastewater in on-going weekly experimentation for COVID-19 surveillance. The novel assay utilized an enzyme capable of reading both DNA and RNA templates, allowing direct amplification of RNA instead of requiring a preceding cDNA synthesis reaction for reverse transcription (Blatter et al., 2013). Molecular targets chosen for analysis included SARS-CoV-2 (targeting the nucleocapsid N3 gene) (Babler et al., 2022), plus indicators of human waste, B2M – specifically the single stranded-mRNA of the protein-coding gene, PMMoV, and SIV. In addition, two quality control targets were analyzed by V2G, OC43 used as the recovery control and HIV used as the inhibition control for this study. The temperature cycling for the V2G assay (for both singleplex and duplex reactions) included an initial denaturation step at 88°C for 30 s., followed by cycling of denaturation occurring at 88 °C for 5 s., an annealing temperature of 60 °C for 20 s., and an elongation/extension temperature of 72 °C for 15 s. The total number of cycles performed per V2G assay was 45×. For the SARS-CoV-2 target, samples were deemed positive if there was amplification by at least one of two replicates plated and run through the reaction.
Master mixes for V2G were hand-crafted (Table 2 ), not via commercial kit, by combining nuclease-free water, V2G reaction buffer (myPOLS LOT# 8016), V2G DNA polymerase (myPOLS LOT# 110521NRA), dNTPs, Platinum Taq antibody (TaKaRa Cat# 9002A), target-specific forward and reverse primers and probes (IDT, Zen – 3’ Iowa Black® Quencher), and an internal passive reference dye, Rox (ThermoFisher Sci. Cat# 12223012), in pre-calculated volumes for a final reaction volume of 40 μL. Two reaction types, with differing reagent volumes were run within this study: 1) for singleplexed targets, SARS-CoV-2 and HIV, and 2) for duplexed targets, PMMoV/SIV, and B2M/OC43 (Table 2). Duplexed reactions included an optimized volume of MgCl2 within the master mix for improving the qPCR efficiency for two targets. All reagents and sample RNA were thawed on ice and remained on ice throughout qPCR setup. A total of 36 μL of master mix was added to each pre-determined well (BIORAD Hard-Shell 96-well PCR plate, #HSP9601), and 4 μL of either sample RNA, nuclease-free water, and target-specific standards with concentrations ranging from 101 to 105 copies/μL (used to develop the standard curve) were added. 96-well plates were loaded with each of the paired experiments RNA samples (A/B: n = 50, C/D: n = 50, E/F: n = 51), seven NTC's per plate for addressing cross-contamination, and one well each of the 101–105 copies/μL standards for quantification. Each plate, once setup, was sealed (BIORAD Microseal® ‘B' Seal, #MSB1001), and briefly centrifuged to remove bubbles from the bottom of each well before it was loaded into the CFX Connect instrument. All qPCR laboratory work occurred within a decontaminated space following standard safety procedures.
Table 2.
Master Mix reagents and volumes for a single-well V2G-qPCR reaction. Sequences of target specific primers and probes utilized per reaction; SARS-CoV-2, and HIV were quantified with the singleplexed 40 μL reaction reagents/ratios below, and B2M/OC43 as well as PMMoV/SIV were quantified with the duplexed 40 uL reaction reagents/ratios.
| V2G-qPCR Master Mix Reagents | Volume per Duplexed 40 µL Reaction |
|---|---|
| Nuclease-free water | 23.31 μL |
| 5× Volcano (2G) Buffer (myPOLS LOT# 8016) | 8.8 μL |
| 10 mM dNTPs | 1.2 μL |
| 5 units/μL Volcano (2G) Polymerase (myPOLS LOT# 110521NRA) | 0.5 μL |
| 5 units/μL Taq Antibody (TaKaRa Cat# 9002A) | 0.25 μL |
| 20 μM forward primer | 0.8 μL |
| 20 μM reverse primer | 0.8 μL |
| 100 μM target-specific FAM probe | 0.16 μL |
| 100 μM target-specific HEX probe | 0.12 μL |
| 400× Rox | 0.1 μL |
| MgCl2 | 0.4 μL |
| V2G-qPCR Master Mix Reagents | Volume per Singleplexed 40 µL Reaction |
|---|---|
| Nuclease-free Water | 23.6 μL |
| 5× Volcano (2G) Buffer (myPOLS LOT# 8016) | 8.8 μL |
| 10 mM dNTPs | 0.8 μL |
| 5 units/μL Volcano (2G) Polymerase (myPOLS LOT# 110521NRA) | 0.4 μL |
| 5 units/μL Taq Antibody (TaKaRa Cat# 9002A) | 0.2 μL |
| 20 μM forward primer | 1 μL |
| 20 μM reverse primer | 1 μL |
| 100 μM target-specific FAM/HEX probe | 0.1 μL |
| 400× Rox | 0.1 μL |
| Molecular Target | Primer/Probe Sequences | |
|---|---|---|
| SARS-CoV-2 (N3) | CV3b/f | TGCTAACAAAGACGGCATCA |
| CV3c/r | GTAGCACGATTGCAGCATTG | |
| CV3.prb | ACA TTG GCA CCC GCA ATC CTG CT (FAM) | |
| SIV | SIV876/f | GCTAGACTCTCACCAGCACTTG |
| SIV999/r | CTAGGAGAGATGGGAACACACA | |
| SIV.prb | TCCACGCTTGCTTGCTTAAAGACCTCT (FAM) | |
| PMMoV | PMMoV/f | AGTGGTTTGACCTTAACGTTTGA |
| PMMoV/r | CCTACGTCTGACGACACAATCT | |
| PMMoV.prb | CCTACCGAAGCAAATGTCGCACT (HEX) | |
| B2M | qB2M/f | CAAGGACTGGTCTTTCTATCTCTTGTAC |
| qB2M/r | CTGCTTACATGTCTCGATCCC | |
| B2M.prb | AGCAGCGGCAGGACCAGCCCCAAG (FAM) | |
| HIV | RTwt3/f | GAAAATTAGTAGATTTCAGAGAACTTAATAAGAGAAC |
| V106/r | CATCACCCACATCCAGTACTGTTA | |
| RT1.prb | TTCTGGGAAGTTCAATTAGGAATACCACATCCCGCAGG (FAM) | |
| OC43 | OC43/f | CAACCAGGCTGATGTCAATAC |
| OC43/r | AAACCTAGTCGGAATAGCCTCA | |
| OC43.prb | ACATTGTCGATCGGGACCCAAGT (HEX) | |
2.7. Data analysis
Shapiro-Wilk normality tests were run on all molecular qPCR datasets plus the measures of FC. Results indicate that the data from this study were generally non-parametric, but a normal distribution was dependent upon molecular target assessed, and sewershed scale of focus. Therefore, Spearman correlation tests were used to evaluate the associations between variables over time. Viral degradation rates were determined, per target, from the slope of the best fit line between the natural logarithm of (Ct/Co) and time, where Co represented the initial target concentration at the first hour measurement interval, and Ct represented the target concentration at hour t (Bamiduro et al., 2021). To compare the results from individual grab samples to the composite sample generated for the degradation set of experiments, the one-sample t-tests were utilized to evaluate whether the mean of the grab samples for the “A” set of experiments was statistically different from the corresponding composite sample. Since the composite was generated from the same 24 grab samples, the use of the mean was deemed more appropriate than the median for the “A” set of experiments given the physical mixing of the grab samples to produce an “average” composite sample.
For the “B” set of experiments, two tests were used for the effective comparison of the 24 grab samples against the composite sample generated, specifically for the SARS-CoV-2 target – one-sample Wilcoxon signed rank tests and one-sample t-tests. The one-sample t-tests were again chosen to compare the means given the physical mixing of the grab samples for the remaining molecular targets, besides SARS-CoV-2. In addition, to determine the level of variability between each sewershed scale from hour-to-hour, homogeneity of variance tests were performed using Levene's tests. All statistical tests performed utilized the statistical software package SPSS version 28.0.0.0. Time series plots were prepared in Microsoft Excel to illustrate the variability of the microbial targets over time. Time series plots for SARS-CoV-2 are shown within the main text (Fig. 3 ) and the plots for the normalized SARS-CoV-2 are shown within the Supplemental text.
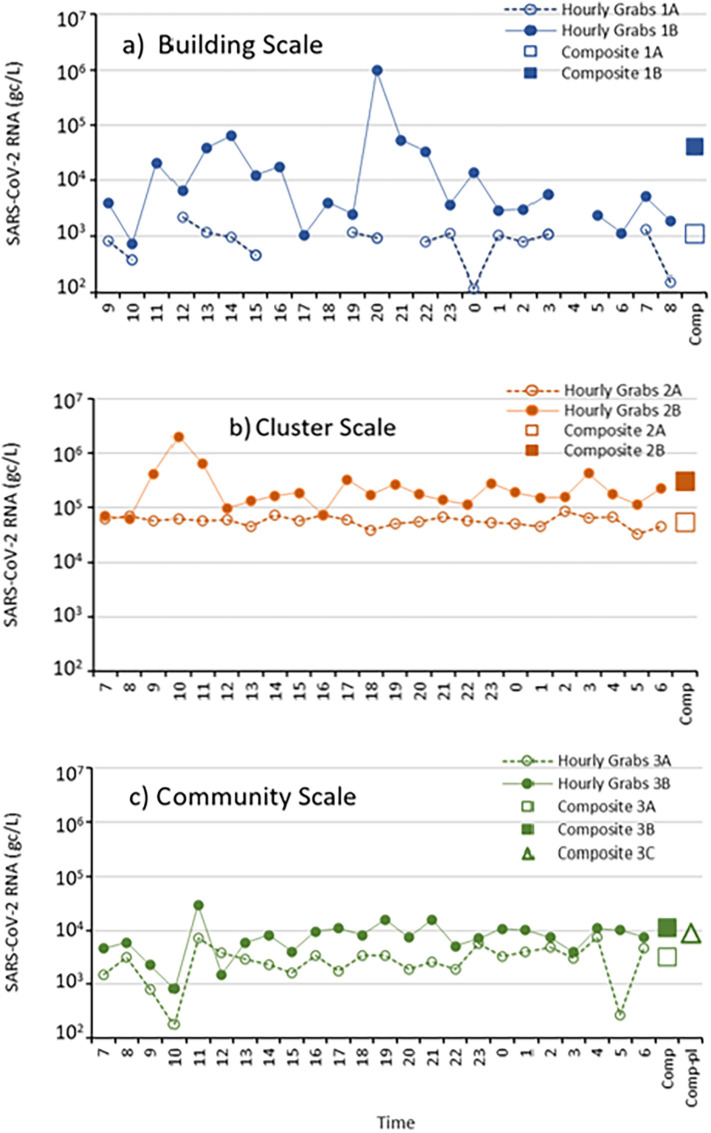
Fig. 3.
Time series plots of SARS-CoV-2 for hourly grab samples and composite samples at the building scale (panel a), cluster scale (panel b) and community scale (panel c). Time shown corresponds to local time the day of the hourly experiment. Composite 3C corresponds to the composite sample collected at the wastewater treatment plant and this sample corresponds to mid-night to mid-night the prior day.
3. Results and discussion
3.1. Degradation of biological signal in wastewater
The persistence of SARS-CoV-2 RNA and its ability to maintain its integrity within the environment for longer than 24 h has been emphasized throughout the literature (Ahmed et al., 2020b; Bivins et al., 2020b; Yang et al., 2022). To provide a comparison of degradation rates, four additional targets (SIV, PMMoV, B2M, and FC) were chosen here, alongside the measurement of SARS-CoV-2, to illustrate the rates at which different biological targets degraded in wastewater (Table 3 ). Each of the molecular targets chosen were comparatively examined to determine if degradation was different at three sewershed scales.
Table 3.
Degradation rates for microbiological targets (hour −1) and Spearman coefficients (r) of study parameters (n = 24 grab samples per experiment, per parameter) over a 24-h time interval from wastewater samples at three distinct sewershed scales. Data for each parameter were transformed from gc/L or CFU/L into ln(Ct/Co) and compared against time (hours). Rows in bold correspond to p<0.05 indicating that the target either increased or decreased significantly over time.
| Experiment: Sewershed Scale | Degradation Rate (k) | Statistical Comparison: ln(Ct/Co) vs. Time (hr) | Spearman's rho (r) | p-value | 95 % Confidence Intervals |
|
|---|---|---|---|---|---|---|
| Lower | Upper | |||||
| Experiment 1A: Building Scale | −0.0265 | SARS-CoV-2 vs. Time | −0.093 | 0.666 | −0.488 | 0.334 |
| −0.0156 | SIV vs. Time | −0.217 | 0.310 | −0.579 | 0.217 | |
| −0.0144 | PMMoV vs. Time | 0.103 | 0.633 | −0.325 | 0.495 | |
| −0.0202 | B2M vs. Time | −0.385 | 0.063 | −0.689 | 0.034 | |
| 0.462 | FC vs. Time | 0.731 | <0.001 | 0.455 | 0.879 | |
| Experiment 2A: Cluster Scale | −0.0102 | SARS-CoV-2 vs. Time | −0.126 | 0.557 | −0.513 | 0.304 |
| −0.0432 | SIV vs. Time | −0.764 | <0.001 | −0.895 | −0.513 | |
| 0.00040 | PMMoV vs. Time | 0.108 | 0.616 | −0.320 | 0.499 | |
| −0.0095 | B2M vs. Time | −0.350 | 0.094 | −0.667 | 0.075 | |
| 0.128 | FC vs. Time | 0.508 | 0.011 | 0.119 | 0.762 | |
| Experiment 3A: Community Scale | 0.0296 | SARS-CoV-2 vs. Time | 0.369 | 0.076 | −0.053 | 0.679 |
| −0.105 | SIV vs. Time | −0.959 | <0.001 | −0.983 | −0.904 | |
| 0.0189 | PMMoV vs. Time | 0.375 | 0.071 | −0.046 | 0.683 | |
| 0.0172 | B2M vs. Time | −0.080 | 0.710 | −0.478 | 0.345 | |
| 0.0218 | FC vs. Time | 0.536 | 0.007 | 0.157 | 0.777 | |
Results from the degradation set of experiments show that at room temperature the RNA of SARS-CoV-2 was stable (Table 3). At all scales (building, cluster, and community) degradation of SARS-CoV-2 RNA was negligible, characterized by low k values (between −0.027 and 0.030 per hour) for collected grab samples. These values are consistent with the lower end of degradation rates as reviewed by McCall et al. (2022). Of interest is that the lower degradation rates corresponded to an exogenous spike of SARS-CoV-2, whereas in the current study our comparatively low degradation rates corresponded to a source endogenous to the wastewater evaluated. In addition results at all scales showed that Spearman correlations for ln(Ct/C0) were not statistically correlated with time (p > 0.07) (Table 3). These results confirm that >24-h are needed for SARS-CoV-2 to degrade to levels that are significantly lower than initial values, which agrees with current literature (Ahmed et al., 2020b; Bivins et al., 2020b; Yang et al., 2022). The study performed by Ahmed et al., 2020b recommended that samples be retained in chilled temperatures, rather than that which is recorded at time of collection. So, through the utilization of ice packs and insulated coolers to transport samples, we agree this handling played a role in reducing the degradation of SARS-CoV-2, endogenous in wastewater, following collection and allowed for quantification of SARS-CoV-2 in a majority of the samples collected by molecular means. Subsequently, the composite sample alone that was created over 24-h, held at room temperature, and analyzed for SARS-CoV-2 resulted in a slight difference of measured RNA from the n = 24 grab average, but resided well within the quantified range of grab samples' viral loads (Table 4 ). Our samples were processed immediately without freezing which may have contributed towards the lack of degradation observed. Bivins et al. (2020b) explains that a limitation in freezing samples is their contribution to viral decay. Samples in the current study were concentrated immediately (< 10 min) upon collection with concentrates refrigerated in DNA/RNA shield for only a few days prior to extraction and qPCR analysis, thereby limiting degradation associated with hold times. t-test results at the cluster (2A) and community (3A) scale further support that SARS-CoV-2 did not degrade significantly (p > 0.393) (Table 4). However, a statistical difference was found between the composite sample and the mean of the 24 grab samples (p < 0.001) at the building scale (1A) (Table 4). This difference was impacted by the fact that the SARS-CoV-2 levels for 8 of the 24 grab samples measured were below detection limits for the building scale experiment. When repeating the t-test with grab samples that measured above detection limits the composite sample mean was no longer statistically different than the average of the grab samples (p = 0.135) (Table 4). A study by Curtis et al. (2021) mentions that grab sample concentrations generally show good agreement with corresponding composite samples for SARS-CoV-2, and that typically only during low incidence or numbers of cases within a community is there a discrepancy. This agrees with our current study in that for the SARS-CoV-2 target there was good representativeness shown between the grab hourly collections and the in-house created composites analyzed by the same methods.
Table 4.
Comparison of microbiological target abundance for 24-h grab sample average versus composite sample produced during the degradation experiments (1-3A). p values of the one-sample t-test <0.05 indicate that the mean of the grab sample was statistically different than the value of the corresponding composite sample.
| Molecular and Microbial Targets | SARS-CoV-2 (gc/L) | SIV (gc/L) | PMMoV (gc/L) | B2M (gc/L) | Fecal coliform by culture (FC) (CFU/L) |
|---|---|---|---|---|---|
| Sample Type | Experiment 1A – Building Scale | ||||
| Grab Average (n = 24) | 6.28 × 102 | 1.93 × 106 | 1.14 × 108 | 1.45 × 106 | 5.33 × 106 |
| Grab Median (n = 24) | 6.14 × 102 | 1.94 × 106 | 1.06 × 108 | 1.48 × 106 | 1.50 × 106 |
| Grab [Min-Max] (Std. Dev) |
[BDL⁎-2.1 × 104] (5.5 × 102) |
[6.3 × 105–3.4 × 106] (4.6 × 105) |
[8.0 × 106–2.7 × 108] (4.4 × 107) | [2.1 × 105–2.2 × 106] (3.8 × 105) | [BDL⁎- 2.8 × 107] (8.0 × 106) |
| Composite 1A | 1.09 × 103 | 1.82 × 106 | 1.32 × 108 | 1.38 × 106 | 1.20 × 107 |
| One-Sample t-test (p-value: two-tailed) | <0.001 ⁎values above detection limits p = 0.135 |
0.284⁎⁎ | 0.050⁎⁎ | 0.365⁎⁎ | <0.001 |
| Sample Type | Experiment 2A – Cluster Scale | ||||
| Grab Average (n = 24) | 5.79 × 104 | 7.12 × 105 | 1.01 × 108 | 3.63 × 105 | 2.31 × 107 |
| Grab Median (n = 24) | 5.83 × 104 | 6.11 × 105 | 1.02 × 108 | 3.62 × 105 | 9.00 × 106 |
| Grab [Min-Max] (Std. Dev) | [3.3 × 104–8.5 × 104] (1.2 × 104) | [3.2 × 105–2.8 × 106] (5.1 × 105) | [8.1 × 107–1.2 × 108] (9.3 × 106) | [2.7 × 105–4.9 × 105] (5.9 × 104) | [1.0 × 103–1.3 × 108] (3.2 × 107) |
| Composite 2A | 5.58 × 104 | 4.03 × 105 | 9.28 × 107 | 3.54 × 105 | 3.40 × 107 |
| One-Sample t-test (p-value: two-tailed) | 0.393⁎⁎ | 0.007 | <0.001 | 0.479⁎⁎ | 0.112⁎⁎ |
| Sample Type | Experiment 3A – Community Scale | ||||
| Grab Average (n = 24) | 3.16 × 103 | 8.92 × 105 | 8.81 × 107 | 1.17 × 105 | 1.13 × 108 |
| Grab Median (n = 24) | 3.11 × 103 | 4.05 × 105 | 9.03 × 107 | 1.18 × 105 | 1.09 × 108 |
| Grab [Min-Max] (Std. Dev) | [1.8 × 102–7.5 × 103] (1.9 × 103) | [2.3 × 105–4.5 × 106] (1.1 × 106) | [3.0 × 107–1.5 × 108] (2.2 × 107) | [7.7 × 103–2.1 × 105] (4.6 × 104) | [6.7 × 107–2.4 × 108] (3.7 × 107) |
| Composite 3A | 3.27 × 103 | 3.09 × 105 | 1.1 × 108 | 1.23 × 105 | 2.21 × 108 |
| One-Sample t-test (p-value: two-tailed) | 0.771⁎⁎ | 0.013 | <0.001 | 0.545⁎⁎ | <0.001 |
BDL = Below Detection Limits. Detection limit for SARS-CoV-2 was 1.0 × 102 gc/L. Detection limit for FC was 1.0 × 106 CFU/L.
Denotes that average of grab and composite sample were statistically not different as per t-test.
Whole SIV particles were spiked-into wastewater as an additional indicator of viral degradation in this study. Degradation was observed for SIV in wastewater collected at the cluster and community sewershed scales, indicating that SIV RNA significantly degraded over 24-h with strong Spearman coefficients (r = −0.764 and r = −0.959 respectively, and p < 0.001) (Table 3). At the building scale, SIV was not recognized to degrade as quickly as at the larger sewersheds, providing no significant correlation with time (r = −0.217, p = 0.31). At the building scale the degradation rate for SIV (k = −0.0156 h−1) measured 4× less than at the cluster scale (k = −0.0432 h−1). At the community scale, SIV degraded roughly 10× more than that of the building scale over the same period (k = −0.105 h−1). This value for SIV degradation is still low in comparison to the high-end measures for SARS-CoV-2 as reviewed by McCall et al. (2022). Overall for SIV, results suggest that at a smaller watershed scale, the RNA of SIV degrades at a slower rate possibly due to the composition of sewage at the small scale in that there are fewer contributing sources and factors impacting the viral particles within the sample. As the sewershed scale increased, the overall degradation of SIV also increased perhaps due to the difference in age and chemistry of the wastewater wherein the sewer system contributed towards a larger variety of different chemicals and organic matter with differing degrading ability towards the virus. To our knowledge, no other studies report on the degradation of SIV within wastewater, so further research is needed to confirm the differences in decay observed here among the sewershed scales. Compared against what was seen with SARS-CoV-2, the t-test results further support that SIV degraded significantly for the cluster and community scale samples as the mean of the grab samples were statistically different than the result from the composite sample (p ≤ 0.013). At the building scale, however, the results from the t-test showed non-statistical differences (p = 0.284) between the mean of the grabs and the value of the composite. As stated above, the chemistry of the building scale wastewater, compared to sewershed scales of larger size, may be an important factor to consider regarding the lack of difference.
PMMoV was another parameter utilized in this study as an indicator of fecal waste and as an additional marker for viral degradation over time as it is a positive-sense single-stranded RNA virus, like that of SARS-CoV-2, found abundantly within wastewater at relatively high concentrations (Zhan et al., 2022; Gerrity et al., 2021; Symonds et al., 2016). Similar to SARS-CoV-2, at all sewershed scales, changes in PMMoV levels had no significant correlation with regards to time (p > 0.07) (Table 3). Furthermore, the degradation coefficients computed were low (k < 0.0189 h−1). Similar to that observed for SARS-CoV-2 RNA, the overall stability of the PMMoV RNA virus measured over subsequent hourly hold times indicates that it takes longer than 24-h for this plant pathogen to degrade to levels that are statistically lower. The limited degradation of PMMoV within differing sewershed scales support its effective use as a normalization parameter within wastewater, as it is a commonly utilized parameter within the literature. Kantor et al. (2021) describe that for normalization to accurately account for SARS-CoV-2 signal loss the degradation rate should remain similar, which coincides with the results of this study for the PMMoV signal. According to the results of the t-tests, there was a statistical difference at cluster and community scales between the mean of the grab samples and that of the composite samples (p < 0.001) (Table 4). At the cluster scale, the composite measured lower than the mean of the grab samples, however at the community scale, the composite sample measured higher than the grab mean. For PMMoV, statistical equivalency was only observed at the building scale (1A; p = 0.050) between both sample types, for the “A” set of experiments. These results suggest that the effective use of PMMoV may depend upon the sewershed scale of focus.
Like SARS-CoV-2 and PMMoV signals, B2M RNA levels were stable with minimal degradation occurring at all scales over 24-h (Table 3). The computed degradation coefficients were relatively low (<0.02 h−1) with insignificant correlations (|r| >0.08, p > 0.06). These results, as observed for SARS-CoV-2 and PMMoV, suggest that B2M takes longer than 24-h to exhibit significantly lower values in wastewater. B2M's composite sample analysis for mRNA abundance had similarly quantified concentrations between the composite sample and average of the grabs, per experiment (Table 4). Consistent with the above, t-test results indicated that for B2M, there was a non-statistical difference in the mean for the grab sample and the value of the composite sample for all sewershed scales (p > 0.365) (Table 4). As compared to PMMoV whose degradation may have been impacted by sewershed scale, the lack of degradation of B2M was observed for all sewershed scales. This consistency may be due to the fact that B2M is a human cellular waste target, rather than that of PMMoV which has a dietary origin and can be variable dependent upon geographic region, sewershed scales, and human behavior. Rather than measurable differences among the sewershed scales between grab and composite samples, like PMMoV, B2M provides a steady signal for normalization purposes (Table 4).
The final target evaluated for degradation rates was FC bacteria, by culture. Results showed evidence of FC multiplication over the 24-h study period at room temperature. The building scale wastewater showed the highest rate of multiplication with an overall positive coefficient (k = 0.462 h−1). As the sewershed size increased, the coefficient declined (Table 3). At the cluster scale the overall rate of multiplication measured at k = 0.128 h−1, and at the community scale it measured at k = 0.0218 h−1. In addition, the building scale had the strongest Spearman correlation (r = 0.731, p ≤0.001), compared to the cluster (r = 0.508, p = 0.011) and community scale (r = 0.536, p = 0.007) (Table 3). At all three sewershed scales, Spearman coefficients were positive and statistically significant providing evidence of multiplication of FC while samples were held at room temperature. As FC bacteria were quantified by CFU's following a period of incubation, it was expected that die-off would occur within the composite sample due to the physical succumbing of bacteria to unknown chemistry of the wastewater samples. However, results showed that the composite sample grew more CFU's than the grabs' mean, and the t-test results indicated that for the building and community scale the mean of the grab sample was statistically different than the value of the composite sample (p < 0.001) (Table 4). A statistical difference between the grab mean, and the composite, was not observed at the cluster scale (p = 0.112).
Overall, degradation was assessed by evaluating both degradation rates and statistical equivalency between the average of grab samples and its corresponding composite. In terms of degradation rates, SARS-CoV-2 appeared to remain stable at all three sewersheds over a 24-h timeframe with PMMoV and B2M behaving in a similar manner, with more stability of B2M across watershed scales. SIV provided a sound surrogate baseline for a degrading signal over time, and as the sewershed scale increased, the more significant the degradation became with regards to the 24-h time interval. In all sewershed scales FC was observed to multiply. It was observed to have multiplied 4-fold from the building to the cluster scale, and 10-fold from the building to the community scale. In terms of statistical equivalency between the average of grab samples and its corresponding composite, consistently across the three sewershed scales, B2M was the only molecular target to have statistically equivalent results between the composite and grab samples, with SARS-CoV-2 following suit showing only a significant difference between the sample types at the building scale. When grab samples below detection were not used in the analysis, SARS-CoV-2 was statistically equivalent at the building scale, thus supporting the overall result showing lack of degradation. Conversely, SIV's measured abundance was only equivalent between the mean of the grabs and the composite sample at the building scale. Mixed results were observed for PMMoV and FC in terms of their statistical differences between the average of the grabs and the composites, as assessed for the “A” set of experiments.
3.2. Hour to hour variability of biological signals in wastewater
Hour to hour variability is expected in wastewater due to the varying uses of water within a building as described by Grijalva et al. (2022). Curtis et al. (2021) provides that multiple parameters could impact the variability of SARS-CoV-2 and other targets concentrations per point in collection which we addressed by measuring the same sampling site consistently each hour for 24 h. In the “B” set of experiments samples were collected every hour from the wastewater source. Parameters which could impact the variability of molecular signal include flow rate (McCall et al., 2022), precipitation or tidal fluctuations (Curtis et al., 2021), and incidence within the community (Bertels et al., 2022). Results from the biological markers show that the average of the 24-h grab samples were within the same order of magnitude of the composite samples for SARS-Cov-2, PMMoV and B2M (Table 5 ). These results are consistent with the results from the “A” set of experiments designed to evaluate degradation rates. As noted above, SARS-CoV-2, PMMoV and B2M did not degrade significantly. Thus, the average of the 24-h grab samples would be expected to represent the composite samples. As mentioned earlier, the stable levels of the endogenous SARS-CoV-2 observed in the current study is consistent with the low end of degradation rates observed in other studies (Ahmed et al., 2020b). Moreover, SIV was shown to degrade during the degradation experiments above. Here for the “B” experiments, the SIV measured from the composite samples were about an order of magnitude lower than that from the average of the 24, hourly grab samples (Table 5). Conversely, the degradation experiments above found evidence of multiplication of FC in the wastewater, and as a result, the composite sample in the “B” set of experiments herein show levels of FC that were higher than the average of the 24-hourly grab samples consistent with the multiplication of FC during the compositing process (Table 5). The extra composite sample, provided for the 3B experiment by the CDWWTP's autosampler, and quantified as a standalone comparative sample, also followed similar orders of magnitude for the molecular and microbial targets of focus; SIV did not degrade as quickly in the WWTP composite sample – measuring one order of magnitude higher than the in-house generated composite. The primary difference between the two composite samples was the slight offset in time of hourly sampling and the temperature of sample storage during compositing. The CDWWTP's autosampler is refrigerated whereas the composite samples prepared manually in the laboratory were held at room temperature. It is possible that the warmer temperature of the laboratory-prepared samples contributed to the degradation of SIV rather than the provided refrigerated sample, which agrees with prior studies that the lower temperatures of sample holding is associated with lower decay rates (Bivins et al., 2020b).
Table 5.
Comparison of microbiological targets for grab versus composite samples produced in the laboratory from samples collected from wastewater source each hour (Experiments 1-3B). p values of the one-sample t-test <0.05 indicate that the mean of the grab sample was statistically different than the value of the corresponding composite sample.
| Molecular and Microbial Targets | SARS-CoV-2 (gc/L) | SIV (gc/L) | PMMoV (gc/L) | B2M (gc/L) | FC (CFU/L) |
|---|---|---|---|---|---|
| Sample Type | Experiment 1B - Building Scale | ||||
| Grab Average (n = 24) | 5.28 × 104 | 3.08 × 106 | 2.43 × 108 | 8.81 × 105 | 2.30 × 107 |
| Grab Median (n = 24) | 4.55 × 103 | 2.32 × 106 | 1.32 × 108 | 8.58 × 105 | 9.00 × 106 |
| Grab [Min-Max](Std. Dev) | [BDL⁎-9.7 × 105] (2.0 × 105) | [1.3 × 105–1.2 × 107] (2.7 × 106) | [6.0 × 107–1.9 × 109] (3.8 × 108) | [5.0 × 103–1.6 × 106] (4.1 × 105) | [BDL⁎-1.9 × 108] (4.0 × 107) |
| Composite 1B | 4.13 × 104 | 1.6 × 105 | 3.70 × 108 | 9.31 × 105 | 1.74 × 108 |
| One-Sample t-test (p-value: two-tailed) | 0.775⁎⁎ | <0.001 | 0.112⁎⁎ | 0.567⁎⁎ | <0.001 |
| Sample Type | Experiment 2B - Cluster Scale | ||||
| Grab Average (n = 24) | 2.82 × 105 | 7.26 × 106 | 1.38 × 108 | 1.08 × 106 | 2.42 × 107 |
| Grab Median (n = 24) | 1.75 × 105 | 6.95 × 106 | 1.40 × 108 | 1.05 × 106 | 1.00 × 107 |
| Grab [Min-Max](Std. Dev) | [6.2 × 104–2.0 × 106] (3.9 × 105) | [3.8 × 106–1.2 × 107] (2.4 × 106) | [5.8 × 107–2.6 × 108] (5.2 × 107) | [4.8 × 105–2.0 × 106] (4.5 × 105) | [1.0 × 103–1.5 × 108] (3.2 × 107) |
| Composite 2B | 3.16 × 105 | 4.38 × 105 | 1.27 × 108 | 9.59 × 105 | 6.80 × 107 |
| One-Sample t-test (p-value: two-tailed) | 0.669⁎⁎ | <0.001 | 0.319⁎⁎ | 0.217⁎⁎ | <0.001 |
| Sample Type | Experiment 3B - Community Scale | ||||
| Grab Average (n = 24) | 8.62 × 103 | 4.30 × 106 | 1.43 × 108 | 2.43 × 105 | 1.02 × 108 |
| Grab Median (n = 24) | 7.63 × 103 | 4.59 × 106 | 1.50 × 108 | 2.61 × 105 | 9.50 × 107 |
| Grab [Min-Max](Std. Dev) | [8.4 × 102–3.0 × 104] (6.1 × 103) | [2.2 × 106–6.5 × 106] (1.1 × 106) | [9.1 × 107–2.1 × 108] (3.2 × 107) | [1.2 × 105–3.6 × 105] (7.0 × 104) | [3.7 × 107–1.7 × 108] (4.0 × 107) |
| Composite 3B | 1.16 × 104 | 6.07 × 105 | 1.66 × 108 | 2.25 × 105 | 4.21 × 108 |
| WWTP Composite | 9.12 × 103 | 6.22 × 106 | 1.78 × 108 | 2.90 × 105 | 2.38 × 108 |
| One-Sample t-test (p-value: two-tailed) | 0.022 (UM) 0.689⁎⁎ (WWTP) |
<0.001 (UM) <0.001 (WWTP) |
0.002 (UM) <0.001 (WWTP) |
0.227⁎⁎ (UM) 0.003 (WWTP) |
<0.001 (UM) <0.001 (WWTP) |
BDL = Below Detection Limits. Detection limit for SARS-CoV-2 was 1.0 × 102 gc/L. Detection limit for FC was 1.0 × 106 CFU/L.
denotes statistical equivalency between grab and composite sample types by Wilcoxon Signed Rank Tests/t-tests.
One-sample t-tests were performed to evaluate the expected representativeness of the hourly collected samples (Exp. “B's”), considering the biological degradation and/or multiplication per target described above, between the grab and composite samples. For SARS-CoV-2, one-sample Wilcoxon signed rank test results showed that there was a statistical difference found between the median of the grab samples and the corresponding value of the composite sample, for all sewershed scales (p < 0.009). These differences were driven by the “spike” nature of the variability of SARS-CoV-2 RNA in samples collected hour to hour, where one hour can comparatively measure much higher or lower than the next. This agrees with the results described by George et al. (2022) in that the representativeness between grab samples with a set frequency of collection and the corresponding composite become reduced when the flow of the wastewater is reduced (Table 1). As observed from the time series plots (Fig. 3), the samples were characterized by orders-of-magnitude variability in some cases. For example, at the cluster scale the majority of the SARS-CoV-2 levels were observed in the 105 gc/L range with the exception of three samples in the 106 gc/L range. These three samples drive up the average SARS-CoV-2 levels in the composite samples (because of elevated RNA levels roughly a factor of 10 above the baseline) but these same samples do not impact the statistical computation of the median because they correspond to a minority of the samples. Therefore, the variable nature of the SARS-CoV-2 signal is best captured by evaluating averages. When using the one-sample t-test, which compares the average of the 24 grab samples in the B set of experiments to the corresponding composite sample, the results illustrate a statistical equivalency between grab and composite sample types for the building and cluster scale (p > 0.669), where the WWTP composite, rather than the in-house created composite, provided equivalency at the community scale (p = 0.689) (Table 5). For all other molecular targets, the results from the comparison of the median of the grabs (one-sample Wilcoxon test) or comparison of the average of the grabs (one-sample t-test) were consistent. Therefore, the discussion from here will focus on emphasizing the results from the one-sample t-test that compares grab against composite sample types.
For the surrogate virus, SIV, consistent with the degradation experiments above, t-test results showed a composite sample average that was less than the average of the 24 grab samples for all sewershed scales and both composite samples at the community scale (p < 0.001) (Table 5). PMMoV and B2M, consistent with the results from the degradation experiments, generally showed statistical equivalency between the average of the grab samples and the composites at each sewershed scale (p > 0.112), with the exception of PMMoV at the community scale where the composite was found to be statistically different from the average of the grab samples (p < 0.002) (Table 5). Given PMMoV's ability to remain relatively stable over 24-h and its widespread usage in wastewater-based epidemiological studies, it was unexpected that PMMoV levels would be significantly different between the sample types at the community scale; the reasons for the exception to the general trend for PMMoV is unknown. Finally, analysis of the FC composite sample, consistent with the results from the degradation experiments, provides evidence that bacterial multiplication is occurring at all three sewershed scales (p < 0.001) (Table 5).
Time series plots of the raw microbial signals show the quantified SARS-CoV-2 RNA (gc/L) from grab samples collected hourly at each sewershed scale (Fig. 3). As grab sampling essentially captured a single point in time of the contributing population to the sewer system, unknown concentrations per target were expected to not only vary over 24-h, but also at the three distinct sewershed scales – as each scale has a distinct servicing population. The building scale was expected to vary the most since the wastewater was collected close to the input source of a small residential population (~500) contributing to the sewer system. The cluster scale was expected to follow a similar trend as that scale encompassed only a few buildings per collection with a roughly 3× greater input population of humans than that of a single building. The community scale, due to the much larger input population (over 1000× greater than a single building), was expected to vary the least overall because the scale servicing hundreds of thousands of individuals essentially averages the microbiome of the sewer system. As observed from the time series plots, hour to hour variability within a sewershed scale can vary by orders of magnitude. At the building scale the results from experiment 1B varied by 3 orders of magnitude (from 103 to 106), and at the cluster scale by 1.5 orders of magnitude (from 105 to 106.5). At the community scale the range for the 3B experiments was also 1.5 orders of magnitude (from 103 to 104.5). Although it appears that the building scale was characterized by more variability, the data should be analyzed statistically to increase confidence in the results and employ the use of averages as the sewershed scale decreases in size.
To evaluate the variability of the SARS-CoV-2 signal between watershed scales, tests of homogeneity of variance were performed on molecular target data, grouped by the sewershed scales. Results showed that the variances between all three sewersheds were statistically equivalent with Levene statistic's ranging from 0.193 to 1.029 (p > 0.455) (Table 6 ). The Levene analyses illustrated that for this study, over a 24-h time interval, the level of variation quantified per molecular target was not different depending on the sewershed scale, contrarily to what was expected. Overall, the variation of each molecular target was not seen to occur between the sewershed scales when they were grouped and assessed as a whole, illustrating that variations over time within a sewershed scale were more impactful than variations observed between the different sewershed scales.
Table 6.
Tests for homogeneity of variance between watershed scales using Levene's Statistics for different molecular and microbial targets, with and without normalization for SARS-CoV-2.
| Molecular/Microbial Target for Experiments (1-3B) | Levene's Test Results |
|
|---|---|---|
| Test statistic | p-value | |
| SARS-CoV-2 | 0.719 | 0.803 |
| SIV | 0.619 | 0.893 |
| PMMoV | 1.029 | 0.455 |
| B2M | 0.193 | 1.000 |
| FC | 0.240 | 1.000 |
| SARS-CoV-2/PMMoV | 0.574 | 0.925 |
| SARS-CoV-2/B2M | 0.966 | 0.521 |
| SARS-CoV-2/FC | 0.770 | 0.748 |
3.2.1. Normalization of SARS-CoV-2 signal
As mentioned above, water sources (toilet water, sink water, shower water, clothes washing water, etc.) of a given sampling location vary over time for downstream applications, and the SARS-CoV-2 signal from WBE applications will be dependent upon the water source (George et al., 2022; Grijalva et al., 2022). To account for the variations in water sources, we utilized three parameters for normalizing the quantified SARS-CoV-2 RNA signal at each sewershed scale (PMMoV, B2M and FC). Previous investigations by Zhan et al. (2022) utilized these three parameters to evaluate the representativeness of human influence for SARS-CoV-2 measurements. Theoretically, normalization by human waste indicators should decrease the hour-to-hour variability of the SARS-CoV-2 signal by removing dilution effects from water sources that have minimal influence from human fluids.
The suitability of normalization was evaluated in two different ways. First the mean of the 24-h grab samples was compared to the composite value, utilizing t-tests, similarly to the raw abundance analyses above, to determine whether normalization improves the ability of the composite samples to represent the 24-h grabs. Second, normalization was evaluated, by comparing the results of the Levene test between the raw (gc/L) SARS-CoV-2 values from the “B” set of experiments against the normalized signal per parameter to determine whether the variability decreased or not.
When SARS-CoV-2 levels were normalized by human waste indicators, the average of 24 grab samples had varied trends of representativeness depending on the fecal parameter used. SARS-CoV-2 normalized by PMMoV was the most consistent across all three sewersheds in that the composite measured well within the range of the grab samples analyzed and was also reported within the same order of magnitude (Table 7 ). t-tests indicated that SARS-CoV-2 data normalized by PMMoV was statistically equivalent between the mean of the 24 grab samples and the composite (p > 0.353) for all three sewershed scales (Table 7). Similarly, SARS-CoV-2 normalized by B2M showed good representativeness between the average of the grabs and composites at all three sewershed scales (Table 7) with most p-values measuring above 0.310 with the exception of the composite for the laboratory prepared composite generated at the community scale which showed the composite (5.2 × 10−2) was higher than the average of the grabs (3.5 × 10−2) (p < 0.001). Finally, for SARS-CoV-2 normalized by FC, there was a larger difference noted between the grab mean and the composites; the normalized composite sample reported roughly 73× lower than the average of the grabs at the building scale, and as the sewershed scale increased the relationship between the sample types became much closer; an 8× decrease was noted for FC normalized SARS-CoV-2 at the cluster scale, and a 3× decrease was seen at the community scale (Table 7). Overall, t-tests supported these differences in that the grab mean and composite sample type did not represent one another and normalization by FC bacteria did not consistently improve the representativeness between the sample types (p < 0.009 with the exception of the building scale). Raw SARS-CoV-2 signals (gc/L), as well as a 3-h moving average of raw SARS-CoV-2, were plotted alongside PMMoV, B2M, and FC normalized SARS-CoV-2 at all three sewersheds over 24-h to provide a visual representation of the hourly variability occurring within the sewer system. These time-series plots can be viewed within the Supplemental Text.
Table 7.
Comparison of normalized SARS-CoV-2 by PMMoV, B2M, and FIB for grab (n = 24) versus composite samples produced in the laboratory from samples collected from the wastewater source each hour (Experiments 1-3B). p values of the one-sample t-test <0.05 indicate that the mean of the grab samples was statistically different than the value of the corresponding composite sample.
| Normalized Molecular and Microbial Targets | SARS-CoV-2/PMMoV | SARS-CoV-2/B2M | SARS-CoV-2/FC |
|---|---|---|---|
| Sample Type | Experiment 1B - Building Scale | ||
| Grab Average (n = 24) | 4.24 × 10−4 | 5.12 × 10−1 | 1.57 |
| Grab Median | 3.26 × 10−5 | 5.75 × 10−3 | 8.28 × 10−4 |
| Grab Range (min – max) | 1.67 × 10−6-7.97 × 10−3 | 3.48 × 10−4-1.08 × 101 | 2.77 × 10−5-2.07 × 101 |
| Grab Std. Dev. | 1.62 × 10−3 | 2.21 | 4.85 |
| Composite 1B | 1.12 × 10−2 | 4.44 × 10−2 | 2.37 × 10−4 |
| One-Sample t-test (p-value: two-tailed) | 0.353⁎⁎ | 0.310⁎⁎ | 0.178⁎⁎ |
| Sample Type | Experiment 2B - Cluster Scale | ||
| Grab Average (n = 24) | 2.10 × 10−3 | 2.85 × 10−1 | 9.88 |
| Grab Median | 1.48 × 10−3 | 1.70 × 10−1 | 1.55 × 10−2 |
| Grab Range (min – max) | 3.83 × 10−4-9.85 × 10−3 | 4.83 × 10−2-1.48 | 1.30 × 10−3-1.65 × 102 |
| Grab Std. Dev. | 2.15 × 10−3 | 3.15 × 10−1 | 3.61 × 101 |
| Composite 2B | 2.49 × 10−3 | 3.29 × 10−1 | 4.65 × 10−3 |
| One-Sample t-test (p-value: two-tailed) | 0.384⁎⁎ | 0.496⁎⁎ | 0.009 |
| Sample Type | Experiment 3B - Community Scale | ||
| Grab Average (n = 24) | 6.41 × 10−5 | 3.45 × 10−2 | 1.00 × 10−4 |
| Grab Median | 4.84 × 10−5 | 3.30 × 10−2 | 7.18 × 10−5 |
| Grab Range (min – max) | 9.32 × 10−6-3.33 × 10−4 | 7.09 × 10−3-1.15 × 10−1 | 1.11 × 10−5-5.02 × 10−4 |
| Grab Std. Dev. | 6.32 × 10−5 | 2.13 × 10−2 | 9.90 × 10−5 |
| Composite 3B | 7.01 × 10−5 | 5.17 × 10−2 | 2.76 × 10−5 |
| WWTP Composite | 5.12 × 10−5 | 3.13 × 10−2 | 3.83 × 10−5 |
| One-Sample t-test (p-value: two-tailed) – Composite 3B | 0.643⁎⁎ | <0.001 | 0.002 |
| One-Sample t-test (p-value: two-tailed) – WWTP Composite | 0.328⁎⁎ | 0.477⁎⁎ | 0.005 |
denotes statistical equivalency between grab and composite sample types by one-sample t-tests.
As mentioned above for SARS-CoV-2, a test for homogeneity of variances was performed on grouped data by sewershed scale providing a Levene statistic of 0.719 (p = 0.803). The null hypothesis for each statistical test assumes equality between the grouped data, so these results were utilized as the baseline comparison for whether the normalization parameters of choice impacted the level of variability seen per sewershed scale. Variability was determined to be reduced if the significance levels (p-values) were closer to one for the normalized SARS-CoV-2 by human waste, and conversely was determined to increase if the significance level approached zero. The mainstream normalization parameter for wastewater-based surveillance studies, PMMoV, was the most impactful over 24-h in reducing the variability of the grouped sewershed scales. Dividing the SARS-CoV-2 abundance by the level of PMMoV RNA detected per hourly collected sample showed that variability decreased, and the equality of the variance increased with a Levene's statistic = 0.574 and more significant p = 0.925 (Table 6). Contrarily, both B2M and FC bacteria did not reduce variability of the SARS-CoV-2 signal over the 24-h time interval in that the Levene test resulted in p-values lower than the stand-alone SARS-CoV-2 measurement (p = 0.521 and p = 0.748, respectively) (Table 6). These results provide that normalization by PMMoV did decrease the variability of the results by lowering the overall variability over time; however, the lowering of the variability was not statistically significant at the different watershed scales.
Overall, the representation between the sampling types for SARS-CoV-2 abundance from grab vs. composite was improved by utilizing normalization. But, given inconsistent findings per target, it was not clear that normalizing SARS-CoV-2 abundance over a 24-h timeframe provided noteworthy benefits, contrarily to what was expected. The viral parameter PMMoV improved the representation of SARS-CoV-2 at the building scale, between the mean of the grabs and composite sample, as well as at the community scale between the grabs and the WWTP provided composite sample (Table 7). Similarly, normalization by B2M improved the representation of SARS-CoV-2 between the sample types at the cluster scale, and between the mean of the grabs and WWTP provided composite (Table 7). Each parameter, as described above, was quantified as relatively stable over 24-h and so provided a consistent value for smoothing out SARS-CoV-2 abundance, supporting the use of these two targets for normalization purposes with WBE studies. FC bacteria did not improve the representativeness between the sample types, but instead worsened the relationship of SARS-CoV-2 abundance estimates per sewershed scale, so is not recommended here as a normalization parameter due to its tendency to replicate over time in the sewer system. From the Levene's test, variability was only observed to decrease through the use of PMMoV as a normalization parameter for SARS-CoV-2 levels in that the central tendencies of the dataset became more equivalent for the same time-period when compared against raw abundance; however this decrease in variability was not statistically significant. B2M levels and FC bacteria were not useful in reducing variability of SARS-CoV-2 when utilized as normalization parameters over 24-h, per sewershed scale, but instead provided more pairwise differences between the sewershed scales upon analysis. Thus overall, normalization of the SARS-CoV-2 signal did not provide consistently improved representations of overall sample means.
3.3. Control targets for quality assurance
The virus OC43 was utilized as a recovery control for this study to provide a baseline comparator for the viral recovery for SARS-CoV-2 following quantification (Kantor et al., 2021; Sharkey et al., 2021; Pecson et al., 2021). The resulting Cq values for the OC43 target per sample are summarized within the Supplemental Text Table S-1, in addition to the calculated percent recovered on a per liter basis. Recoveries ranged from 13 to 33 % for the grab samples and 3 to 38 % for the composite samples. To note, within this study reported values for molecular targets (including SARS-CoV-2) are direct measures and were therefore not adjusted for recovery of OC43. We recommend that the degradation of OC43 should be considered when calculating % recovery as the composite sample had less OC43 recovered overall (except Exp B. for the UM composite) compared against the 24-h grab average which were processed immediately, indicating degradation for this target. Holding time of samples at room temperature could be a factor in this study that provided lower % recoveries for OC43 within the composite samples. Further research is needed regarding the decay rate of this commonly utilized processing control.
Comparison of the Cq values of the HIV inhibition control against a water-RNA control (30 μL of HIV RNA + 10 μL nuclease-free water) showed little to no inhibition of wastewater collected at all three sewershed scales. More details about the inhibition analysis by the HIV target is available in the Supplemental Text.
4. Conclusions
The use of WBE as a tool for predicting the prevalence of COVID-10 within communities is increasing in popularity and scope, but work has led to a better understanding of its limitations for its predictive capacity and power as a tool for guiding public health policies (Haramoto et al., 2020; Sherchan et al., 2020; Westhaus et al., 2021; Kitajima et al., 2020). In this study we addressed the representativeness of composite sampling types, made observations about biological degradation occurring within wastewater using multiple microbial targets, and assessed variability occurring intra-day and from hour to hour while evaluating the potential of normalization parameters of human waste. This study is unique in that it comprehensively evaluated degradation and hour to hour variability, using both endogenous and exogenous microbial targets while monitoring for basic physical chemical parameters. Degradation experiments were conducted simultaneously with hour-to-hour wastewater variability studies allowing for the assessment of sample representativeness using the same wastewater sample, thereby controlling for wastewater chemistry.
Overall, lack of significant degradation of SARS-CoV-2, PMMoV, and B2M suggests that composite samples are not adversely affected by the 24-h sample hold time within the automatic composite sampler. So composite sampling is recommended for measurements of SARS-CoV-2, PMMoV, and B2M due to their relative stability over a period of 24 h. Specifically for SARS-CoV-2, degradation was insignificant. This differs from other studies which generally indicate measurable degradation (McCall et al., 2022). We believe that the lack of degradation in the current study was due to the measurement of endogenous SARS-CoV-2, PMMoV, and B2M coupled with the characteristics of the wastewater analyzed, and the immediate concentration and stabilization of sample DNA/RNA. We also found that the degradation of the exogenous virus, SIV, was a function of sewershed scale with no significant degradation observed at the building scale in contrast to the significant degradation observed at the cluster and community scale.
In this study we found that the microbial targets varied from one hour to the next within a 24-h timeframe, so the composite sample reflected the average of the SARS-CoV-2 signal from individual grabs. However, significant degradation of SIV, and multiplication of FC bacteria showed that for some biological targets, composite sampling may impact the level of abundance quantified downstream, so results should be corrected for die-off or growth. The degradation seen here for SIV may have been offset using refrigeration during the compositing processes, but more research is needed to confirm the importance of temperature for this viral target. Studies aimed at collecting and evaluating other microbiological targets from wastewater should evaluate the need for refrigeration as this study suggests that rate of decay is different between targets found within wastewater.
Normalization by common, and less-common, indicators of human waste was also a strategy utilized here to improve the representativeness between grab and composite sampling for SARS-CoV-2. Although some reductions in variability were observed when the SARS-CoV-2 was normalized by PMMoV, the overall improvements made with normalization between sampling types were considered minimal.
In cases where composite samples are not feasible, we recommend grab sampling over the long term at the same time of day and day of week to reduce variability caused by changes in water use. This recommendation is consistent with those of Grijalva et al. (2022) who emphasized that the daily variability of SARS-CoV-2 signals within wastewater has yet to be leveraged for best-practices of collecting wastewater grabs. In addition to evaluating utility of daily sampling at the same time of day, we recommend that future experiments augment the work from current studies (Ahmed et al., 2021; George et al., 2022) by collecting samples every hour for a period of a week, or at shorter time-frequencies of collection (i.e., every 10–15 min) over 24-h, to evaluate across study consistency in sample timing to control for variability caused by changes in water use.
CRediT authorship contribution statement
Krstina M. Babler: Conceptualization, Methodology, Formal Analysis, Writing – Original Draft. Mark E. Sharkey: Conceptualization, Methodology, Formal Analysis, Writing-Review. Samantha Abelson: Methodology. Ayaaz Amirali: Methodology. Aymara Benitez: Resources. Gabriella A. Cosculluela: Methodology. George S. Grills: Conceptualization, Writing – Review. Naresh Kumar: Conceptualization, Methodology. Jennifer Laine: Methodology. Walter Lamar: Methodology. Erik Lamm: Methodology. Jiangnan Lyu: Formal Analysis. Christopher Mason: Conceptualization, Funding Acquisition. Philip M McCabe: Resources. Joshi Raghavender: Resources. Brian D. Reding: Methodology. Matthew A. Roca: Methodology. Stephan C. Schurer: Data Standardization, Funding Acquisition. Mario Stevenson: Resources. Angela Szeto: Resources. John J. Tallon Jr.: Methodology, Resources. Dusica Vidovic: Data Standardization. Yalda Zarnegarnia: Formal Analysis. Helena M. Solo-Gabriele: Conceptualization, Methodology, Visualization, Formal Analysis, Writing – Review, Supervision, Project Administration, Funding Acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This project is supported by the National Institutes of Health (NIH) Rapid Acceleration of Diagnostics - Radical (RADx-rad) grant 1U01DA053941; the NIH / NCI Cancer Center Support Grant 1P30CA240139-01 for the Sylvester Comprehensive Cancer Center; NIH grant P30AI073961 for the Miami Center for AIDS Research (CFAR); and NIH grant 1UL1TR000460 for the University of Miami Clinical and Translational Science Institute (CTSI). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. This project is also supported by University of Miami internal funding for wastewater surveillance at the university. We are grateful to the support from the University of Miami Facilities Administration and to the laboratory personnel from the Miami-Dade Water and Sewer Department who provided considerable logistical support throughout the 24-h sampling and analysis efforts. Dr. Chris Mason was also supported by Testing for America (501c3), OpenCovidScreen Foundation, the Bert L and N Kuggie Vallee Foundation, Igor Tulchinsky and the WorldQuant Foundation, Bill Ackman and Olivia Flatto and the Pershing Square Foundation, Ken Griffin and Citadel, the US National Institutes of Health (R01AI125416, R21AI129851, R01AI151059, U01DA053941), the Rockefeller Foundation, and the Alfred P. Sloan Foundation (G-2015-13964).
Editor: Warish Ahmed
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2023.161423.
Appendix A. Supplementary data
Supplementary material for items referenced in the manuscript inclusive of water quality results, summary of OC43 control, summary of HIV control, and normalization of SARS-CoV-2 by three parameters.
Data availability
Data will be made available on request.
References
- Abdelzaher A.M., Solo-Gabriele H.M., Wright M.E., Palmer C.J. Sequential concentration of bacteria and viruses from marine waters using a dual membrane system. J. Environ. Qual. 2008;37(4):1648–1655. doi: 10.2134/jeq2007.0238. [DOI] [PubMed] [Google Scholar]
- Abdelzaher A.M., Solo-Gabriele H.M., Palmer C.J., Scott T.M. Simultaneous concentration of Enterococci and coliphage from marine waters using a dual layer filtration system. J. Environ. Qual. 2009;38(6):2468–2473. doi: 10.2134/jeq2008.0488. [DOI] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bibby K., Haramoto E., Hewitt J., Huygens F., Gyawali P., Korajkic A., Riddell S., Sherchan S.P., Simpson S.L., Sirikanchana K., Symonds E.M., Verhagen R., Vasan S.S., Kitajima M., Bivins A. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020;191 doi: 10.1016/j.envres.2020.110092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bivins A., Bertsch P.M., Bibby K., Choi P.M., Farkas K., Gyawali P., Hamilton K.A., Haramoto E., Kitajima M., Simpson S.L., Tandukar S., Thomas K., Mueller J.F. Surveillance of SARS-CoV-2 RNA in wastewater: methods optimisation and quality control are crucial for generating reliable public health information. Curr. Opin. Environ. Sci. Health. 2020;17:82–93. doi: 10.1016/j.coesh.2020.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bivins A., Bertsch P.M., Bibby K., Gyawali P., Sherchan S.P., Simpson S.L., Thomas K.V., Verhagen R., Kitajima M., Mueller J.F., Korajkic A. Intraday variability of indicator and pathogenic viruses in 1-h and 24-h composite wastewater samples: implications for wastewater-based epidemiology. Environ. Res. 2021;193 doi: 10.1016/j.envres.2020.110531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Simpson S.L., Bertsch P.M., Bibby K., Bivins A., Blackall L.L., Bofill-Mas S., Bosch A., Brandao J., Choi P.M., Ciesielski M., Donner E., D'Souza N., Farnleitner A.H., Gerrity D., Gonzalez R., Griffith J.F., Gyawali P., Haas C.N., Hamilton K.A., Hapuarachchi H.C., Harwood V.J., Haque R., Jackson G., Khan S.J., Khan W., Kitajima M., Korajkic A., La Rosa G., Layton B.A., Lipp E., McLellan S.L., McMinn B., Medema G., Metcalfe S., Meijer W.G., Mueller J.F., Murphy H., Naughton C.C., Noble R.T., Payyappat S., Petterson S., Pitkanen T., Rajal V.B., Reyneke B., Roman F.A., Jr., Rose J.B., Rusinol M., Sadowsky M.J., Sala-Comorera L., Setoh Y.X., Sherchan S.P., Sirikanchana K., Smith W., Steele J.A., Sabburg R., Symonds E.M., Thai P., Thomas K.V., Tynan J., Toze S., Thompson J., Whiteley A.S., Wong J.C.C., Sano D., Wuertz S., Xagoraraki I., Zhang Q., Zimmer-Faust A.G., Shanks O.C. Minimizing errors in RT-PCR detection and quantification of SARS-CoV-2 RNA for wastewater surveillance. Sci. Total Environ. 2022;805 doi: 10.1016/j.scitotenv.2021.149877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- APHA, WEF, AWWA . 21st edition. 2005. Standard Methods for the Examination of Water and Wastewater. [Google Scholar]
- APHA, AWWA, WEF . 23rd edition. 2017. Standard Methods for the Examination of Water and Wastewater. 9510 Detection of Enteric Viruses. [Google Scholar]
- Babler K.M., Amirali A., Sharkey M.E., Williams S.L., Boone M.M., Cosculluela G.A., Currall B.B., Grills G.S., Laine J., Mason C.E., Reding B.D., Schurer S.C., Stevenson M., Vidovic D., Solo-Gabriele H.M. Comparison of electronegative filtration to magnetic bead-based concentration and V2G-qPCR to RT-qPCR for quantifying viral SARS-CoV-2 RNA from wastewater. ACS ES&T-Water. 2022;2(11):2004–2013. doi: 10.1021/acsestwater.2c00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamiduro G.J., Kumar N., Solo-Gabriele H.M., Zahran E.M. Persistence of aerially-sprayed naled in coastal sediments. Sci. Total Environ. 2021;794 doi: 10.1016/j.scitotenv.2021.148701. [DOI] [PubMed] [Google Scholar]
- Bertanza G., Boiocchi R., Pedrazzani R. Improving the quality of wastewater treatment plant monitoring by adopting proper sampling strategies and data processing criteria. Sci. Total Environ. 2022;806(Pt 3) doi: 10.1016/j.scitotenv.2021.150724. [DOI] [PubMed] [Google Scholar]
- Bertels X., Demeyer P., Van den Bogaert S., Boogaerts T., van Nuijs A.L.N., Delputte P., Lahousse L. Factors influencing SARS-CoV-2 RNA concentrations in wastewater up to the sampling stage: a systematic review. Sci. Total Environ. 2022;820 doi: 10.1016/j.scitotenv.2022.153290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt W.Q., Schmitz B.W., Innes G.K., Prasek S.M., Pogreba Brown K.M., Stark E.R., Foster A.R., Sprissler R.S., Harris D.T., Sherchan S.P., Gerba C.P., Pepper I.L. COVID-19 containment on a college campus via wastewater-based epidemiology, targeted clinical testing and an intervention. Sci. Total Environ. 2021;779 doi: 10.1016/j.scitotenv.2021.146408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., North D., Ahmad A., Ahmed W., Alm E., Been F., Bhattacharya P., Bijlsma L., Boehm A.B., Brown J., Buttiglieri G., Calabro V., Carducci A., Castiglioni S., Cetecioglu Gurol Z., Chakraborty S., Costa F., Curcio S., de Los Reyes F.L., III, Delgado Vela J., Farkas K., Fernandez-Casi X., Gerba C., Gerrity D., Girones R., Gonzalez R., Haramoto E., Harris A., Holden P.A., Islam M.T., Jones D.L., Kasprzyk-Hordern B., Kitajima M., Kotlarz N., Kumar M., Kuroda K., La Rosa G., Malpei F., Mautus M., McLellan S.L., Medema G., Meschke J.S., Mueller J., Newton R.J., Nilsson D., Noble R.T., van Nuijs A., Peccia J., Perkins T.A., Pickering A.J., Rose J., Sanchez G., Smith A., Stadler L., Stauber C., Thomas K., van der Voorn T., Wigginton K., Zhu K., Bibby K. Wastewater-based epidemiology: global collaborative to maximize contributions in the fight against COVID-19. Environ. Sci. Technol. 2020;54(13):7754–7757. doi: 10.1021/acs.est.0c02388. [DOI] [PubMed] [Google Scholar]
- Bivins A., Greaves J., Fischer R., Yinda K.C., Ahmed W., Kitajima M., Munster V.J., Bibby K. Persistence of SARS-CoV-2 in water and wastewater. Environ.Sci.Technol.Lett. 2020;7:937–942. doi: 10.1021/acs.estlett.0c00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatter N., Bergen K., Nolte O., Welte W., Diederichs K., Mayer J., Wieland M., Marx A. Structure and function of an RNA-reading thermostable DNA polymerase. Angew. Chem. Int. Ed. Engl. 2013;52(45):11935–11939. doi: 10.1002/anie.201306655. [DOI] [PubMed] [Google Scholar]
- Curtis K., Keeling D., Yetka K., Larson A., Gonzalez R. 2021. Wastewater SARS-CoV-2 RNA Concentration and Loading Variability From Grab and 24-Hour Composite Samples. medRxiv. [Google Scholar]
- Davoodi R., Pirsaheb M., Karimyan K., Gupta V.K., Takhtshahi A.R., Sharafi H., Moradi M. Data for distribution of various species of fecal coliforms in urban, rural and private drinking water sources in ten years period - a case study: Kermanshah,Iran. Data Brief. 2018;18:1544–1550. doi: 10.1016/j.dib.2018.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George A.D., Kaya D., Layton B.A., Bailey K., Mansell S., Kelly C., Williamson K.J., Radniecki T.S. Impact of sampling type, frequency, and scale of the collection system on SARS-CoV-2 quantification fidelity. Environ.Sci.Technol.Lett. 2022;9:160–165. doi: 10.1021/acs.estlett.1c00882. [DOI] [PubMed] [Google Scholar]
- Gerrity D., Papp K., Stoker M., Sims A., Frehner W. Early-pandemic wastewater surveillance of SARS-CoV-2 in Southern Nevada: methodology, occurrence, and incidence/prevalence considerations. Water Res. X. 2021;10 doi: 10.1016/j.wroa.2020.100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibas C., Lambirth K., Mittal N., Juel M.A.I., Barua V.B., Roppolo Brazell L., Hinton K., Lontai J., Stark N., Young I., Quach C., Russ M., Kauer J., Nicolosi B., Chen D., Akella S., Tang W., Schlueter J., Munir M. Implementing building-level SARS-CoV-2 wastewater surveillance on a university campus. Sci. Total Environ. 2021;782 doi: 10.1016/j.scitotenv.2021.146749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grijalva L.M., Brown B., Cauble A., Tarpeh W.A. Diurnal variability of SARS-CoV-2 RNA concentrations in hourly grab samples of wastewater influent during low COVID-19 incidence. ACS ES&T Water. 2022;2(11):2125–2133. doi: 10.1021/acsestwater.2c00061. [DOI] [PubMed] [Google Scholar]
- Gussow D., Rein R., Ginjaar I., Hochstenbach F., Seemann G., Kottman A., Ploegh H.L. The human beta 2-microglobulin gene. Primary structure and definition of the transcriptional unit. J. Immunol. 1987;139(9):3132–3138. [PubMed] [Google Scholar]
- Hachich E.M., Di Bari M., Christ A.P., Lamparelli C.C., Ramos S.S., Sato M.I. Comparison of thermotolerant coliforms and Escherichia coli densities in freshwater bodies. Braz. J. Microbiol. 2012;43(2):675–681. doi: 10.1590/S1517-83822012000200032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramoto E., Malla B., Thakali O., Kitajima M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 2020;737 doi: 10.1016/j.scitotenv.2020.140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart O.E., Halden R.U. Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: feasibility, economy, opportunities and challenges. Sci. Total Environ. 2020;730 doi: 10.1016/j.scitotenv.2020.138875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juel M.A.I., Stark N., Nicolosi B., Lontai J., Lambirth K., Schlueter J., Gibas C., Munir M. Performance evaluation of virus concentration methods for implementing SARS-CoV-2 wastewater based epidemiology emphasizing quick data turnaround. Sci. Total Environ. 2021;801 doi: 10.1016/j.scitotenv.2021.149656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantor R.S., Nelson K.L., Greenwald H.D., Kennedy L.C. Challenges in measuring the recovery of SARS-CoV-2 from wastewater. Environ. Sci. Technol. 2021;55(6):3514–3519. doi: 10.1021/acs.est.0c08210. [DOI] [PubMed] [Google Scholar]
- Kim S., Kennedy L.C., Wolfe M.K., Criddle C.S., Duong D.H., Topol A., White B.J., Kantor R.S., Nelson K.L., Steele J.A., Langlois K., Griffith J.F., Zimmer-Faust A.G., McLellan S.L., Schussman M.K., Ammerman M., Wigginton K.R., Bakker K.M., Boehm A.B. SARS-CoV-2 RNA is enriched by orders of magnitude in primary settled solids relative to liquid wastewater at publicly owned treatment works. Environ.Sci. (Camb.) 2022;8(4):757–770. doi: 10.1039/d1ew00826a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M., Ahmed W., Bibby K., Carducci A., Gerba C.P., Hamilton K.A., Haramoto E., Rose J.B. SARS-CoV-2 in wastewater: state of the knowledge and research needs. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhang S., Shi J., Luby S.P., Jiang G. Uncertainties in estimating SARS-CoV-2 prevalence by wastewater-based epidemiology. Chem. Eng. J. 2021;415 doi: 10.1016/j.cej.2021.129039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmud Z.H., Islam M.S., Imran K.M., Hakim S.A.I., Worth M., Ahmed A., Hossan S., Haider M., Islam M.R., Hossain F., Johnston D., Ahmed N. Occurrence of Escherichia coli and faecal coliforms in drinking water at source and household point-of-use in Rohingya camps,Bangladesh. Gut Pathog. 2019;11:52. doi: 10.1186/s13099-019-0333-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall C., Fang Z.N., Li D., Czubai A.J., Juan A., LaTurner Z.W., Ensor K., Hopkins L., Bedient P.B., Stadler L.B. Modeling SARS-CoV-2 RNA degradation in small and large sewersheds. Environ.Sci.Water Res.Technol. 2022:8. [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M., Warren J.L., Weinberger D.M., Arnold W., Omer S.B. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38(10):1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecson B.M., Darby E., Haas C.N., Amha Y.M., Bartolo M., Danielson R., Dearborn Y., Di Giovanni G., Ferguson C., Fevig S., Gaddis E., Gray D., Lukasik G., Mull B., Olivas L., Olivieri A., Qu Y., Consortium S.A.-C.-I. Reproducibility and sensitivity of 36 methods to quantify the SARS-CoV-2 genetic signal in raw wastewater: findings from an interlaboratory methods evaluation in the U.S. Environ. Sci. (Camb.) 2021;7:504–520. doi: 10.1039/d0ew00946f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philo S.E., Keim E.K., Swanstrom R., Ong A.Q.W., Burnor E.A., Kossik A.L., Harrison J.C., Demeke B.A., Zhou N.A., Beck N.K., Shirai J.H., Meschke J.S. A comparison of SARS-CoV-2 wastewater concentration methods for environmental surveillance. Sci. Total Environ. 2021;760 doi: 10.1016/j.scitotenv.2020.144215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philo S.E., Ong A.Q.W., Keim E.K., Swanstrom R., Kossik A.L., Zhou N.A., Beck N.K., Meschke J.S. Development and validation of the skimmed milk pellet extraction protocol for SARS-CoV-2 wastewater surveillance. Food Environ. Virol. 2022;14:355–363. doi: 10.1007/s12560-022-09512-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Cuevas-Ferrando E., Sanjuan R., Domingo-Calap P., Sanchez G. Metropolitan wastewater analysis for COVID-19 epidemiological surveillance. Int. J. Hyg. Environ. Health. 2020;230 doi: 10.1016/j.ijheh.2020.113621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosiles-Gonzalez G., Carrillo-Jovel V.H., Alzate-Gaviria L., Betancourt W.Q., Gerba C.P., Moreno-Valenzuela O.A., Tapia-Tussell R., Hernandez-Zepeda C. Environmental surveillance of SARS-CoV-2 RNA in wastewater and groundwater in Quintana Roo,Mexico. Food Environ. Virol. 2021;13(4):457–469. doi: 10.1007/s12560-021-09492-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz B.W., Innes G.K., Prasek S.M., Betancourt W.Q., Stark E.R., Foster A.R., Abraham A.G., Gerba C.P., Pepper I.L. Enumerating asymptomatic COVID-19 cases and estimating SARS-CoV-2 fecal shedding rates via wastewater-based epidemiology. Sci. Total Environ. 2021;801 doi: 10.1016/j.scitotenv.2021.149794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schussman M.K., McLellan S.L. Effect of time and temperature on SARS-CoV-2 in municipal wastewater conveyance systems. Water (Basel) 2022:14. [Google Scholar]
- Sharkey M.E., Kumar N., Mantero A.M.A., Babler K.M., Boone M.M., Cardentey Y., Cortizas E.M., Grills G.S., Herrin J., Kemper J.M., Kenney R., Kobetz E., Laine J., Lamar W.E., Mader C.C., Mason C.E., Quintero A.Z., Reding B.D., Roca M.A., Ryon K., Solle N.S., Schurer S.C., Shukla B., Stevenson M., Stone T., Tallon J.J., Jr., Venkatapuram S.S., Vidovic D., Williams S.L., Young B., Solo-Gabriele H.M. Lessons learned from SARS-CoV-2 measurements in wastewater. Sci. Total Environ. 2021;798 doi: 10.1016/j.scitotenv.2021.149177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherchan S.P., Shahin S., Ward L.M., Tandukar S., Aw T.G., Schmitz B., Ahmed W., Kitajima M. First detection of SARS-CoV-2 RNA in wastewater in North America: a study in Louisiana, USA. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solo-Gabriele H.M., Kumar S., Abelson S., Penso J., Contreras J., Babler K.M., Sharkey M.E., Mantero A.M.A., Lamar W.E., Tallon J.J., Kobetz E., Solle N.S., Shukla B.S., Kenney R.J., Mason C.E., Schurer S.C., Vidovic D., Williams S.L., Grills G.S., Jayaweera D.T., Mirsaeidi M., Kumar N. 2022. COVID-19 Prediction using Genomic Footprint of SARS-CoV-2 in Air, Surface Swab and Wastewater Samples. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z., Reinke R., Hoxsey M., Jackson J., EKrikorian E., Melitas N., Rosso D., Jiang S. Detection of SARS-CoV-2 in wastewater: community variability, temporal dynamics, and genotype diversity. ACS ES&T Water. 2021;1:1816–1825. [Google Scholar]
- Symonds E.M., Sinigalliano C., Gidley M., Ahmed W., McQuaig-Ulrich S.M., Breitbart M. Faecal pollution along the southeastern coast of Florida and insight into the use of pepper mild mottle virus as an indicator. J. Appl. Microbiol. 2016;121(5):1469–1481. doi: 10.1111/jam.13252. [DOI] [PubMed] [Google Scholar]
- Vo V., Tillett R.L., Chang C.L., Gerrity D., Betancourt W.Q., Oh E.C. SARS-CoV-2 variant detection at a university dormitory using wastewater genomic tools. Sci. Total Environ. 2022;805 doi: 10.1016/j.scitotenv.2021.149930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidhaas J., Aanderud Z.T., Roper D.K., VanDerslice J., Gaddis E.B., Ostermiller J., Hoffman K., Jamal R., Heck P., Zhang Y., Torgersen K., Laan J.V., LaCross N. Correlation of SARS-CoV-2 RNA in wastewater with COVID-19 disease burden in sewersheds. Sci. Total Environ. 2021;775 doi: 10.1016/j.scitotenv.2021.145790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhaus S., Weber F.A., Schiwy S., Linnemann V., Brinkmann M., Widera M., Greve C., Janke A., Hollert H., Wintgens T., Ciesek S. Detection of SARS-CoV-2 in raw and treated wastewater in Germany - suitability for COVID-19 surveillance and potential transmission risks. Sci. Total Environ. 2021;751 doi: 10.1016/j.scitotenv.2020.141750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilder M.L., Middleton F., Larsen D.A., Du Q., Fenty A., Zeng T., Insaf T., Kilaru P., Collins M., Kmush B., Green H.C. Co-quantification of crAssphage increases confidence in wastewater-based epidemiology for SARS-CoV-2 in low prevalence areas. Water Res. X. 2021;11 doi: 10.1016/j.wroa.2021.100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Xiao A., Zhang J., Moniz K., Endo N., Armas F., Bonneau R., Brown M.A., Bushman M., Chai P.R., Duvallet C., Erickson T.B., Foppe K., Ghaeli N., Gu X., Hanage W.P., Huang K.H., Lee W.L., Matus M., McElroy K.A., Nagler J., Rhode S.F., Santillana M., Tucker J.A., Wuertz S., Zhao S., Thompson J., Alm E. SARS-CoV-2 RNA concentrations in wastewater foreshadow dynamics and clinical presentations of new COVID-19 cases. Sci. Total Environ. 2022;805 doi: 10.1016/j.scitotenv.2021.150121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Dong Q., Li S., Cheng Z., Kang X., Ren D., Xu C., Zhou X., Liang P., Sun L., Zhao J., Jiao Y., Han T., Liu Y., Qian Y., Liu Y., Huang X., Qu J. Persistence of SARS-CoV-2 RNA in wastewater after the end of the COVID-19 epidemics. J. Hazard. Mater. 2022;429 doi: 10.1016/j.jhazmat.2022.128358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan Q., Babler K.M., Sharkey M.E., Amirali A., Beaver C.C., Boone M.M., Comerford S., Cooper D., Cortizas E.M., Currall B.B., Foox J., Grills G.S., Kobetz E., Kumar N., Laine J., Lamar W.E., Mantero A.M.A., Mason C.E., Reding B.D., Robertson M., Roca M.A., Ryon K., Schurer S.C., Shukla B.S., Solle N.S., Stevenson M., Tallon J.J.J., Thomas C., Thomas T., Vidovic D., Williams S.L., Yin X., Solo-Gabriele H.M. Relationships between SARS-CoV-2 in wastewater and COVID-19 clinical cases and hospitalizations, with and without normalization against indicators of human waste. ACS ES&T-Water. 2022;2(11):1992–2003. doi: 10.1021/acsestwater.2c00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Chen L., Shen Z. Impacts of rapid urbanization on characteristics, sources and variation of fecal coliform at watershed scale. J. Environ. Manag. 2021;286 doi: 10.1016/j.jenvman.2021.112195. [DOI] [PubMed] [Google Scholar]
- Zulli A., Pan A., Bart S.M., Crawford F.W., Kaplan E.H., Cartter M., Ko A.I., Sanchez M., Brown C., Cozens D., Brackney D.E., Peccia J. Predicting daily COVID-19 case rates from SARS-CoV-2 RNA concentrations across a diversity of wastewater catchments. FEMS Microbes. 2021;2 doi: 10.1093/femsmc/xtab022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for items referenced in the manuscript inclusive of water quality results, summary of OC43 control, summary of HIV control, and normalization of SARS-CoV-2 by three parameters.
Data Availability Statement
Data will be made available on request.