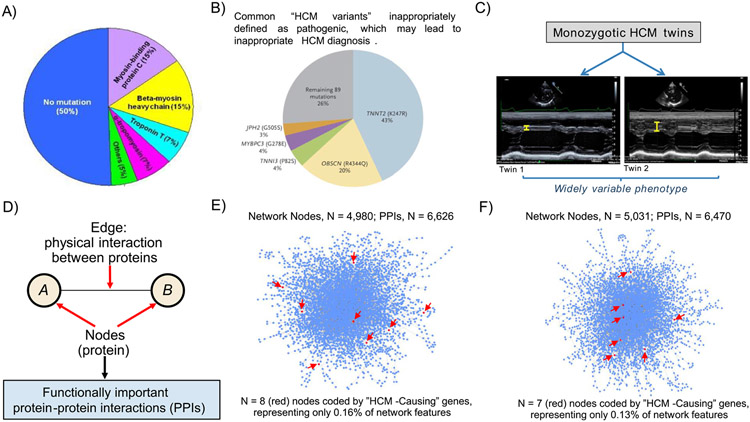
Figure 1.
Determinants of HCM. (A) Distribution of variants in genes coding for sarcomere proteins in patients with HCM assessed in clinical practice. It is notable that 50% of HCM patients have no sarcomere gene mutation. However, only 30% of consecutive HCM patients have mutations that can be considered pathogenic (disease-causing). (B) Prevalence of putative HCM-causing gene variants is greater than expected in large genetic databases comprising non-affected populations: TNNT2 (K247R), OBSCN (R4344Q), TNNI3 (P82S), MYBPC3 (G278E), and JPH2 (G505S) are top 5 most common gene variants inappropriately defined as pathogenic, and account for most (74%) of HCM diagnoses judged misclassified. Reproduced with permission from Manrai et al.15 (C) Recent data from the Seidman laboratory shows widely divergent HCM phenotypes in monozygotic twins, supporting the hypothesis that factors other than genotype are critical to understanding the totality of HCM. Ventricular septal thickness on echocardiographic imaging differs substantially in the twins. Reproduced from Repetli GG, et al28 with permission. (D) Network features, emphasizing that network connections represent functionally important protein-protein interactions (PPIs). (E, F) Using myectomy specimens from patients with obstructive HCM, PPI networks are dense, complex, and vary widely between patients,31 suggesting that sarcomere genes alone are unlikely to explain the pathogenesis of this disease. Network topology was not informed by putative HCM-causing sarcomere genes, which in fact represented only <0.1% of all network features. Two representative patient networks are shown, which underscore: (i) HCM pathobiology is complex and (ii) a vast majority of functionally important molecular signaling pathways do not involve any of 17 genes commonly designated as HCM disease-causing (red arrows). Images derived from methods and results reported originally in Maron et al.31Heterogeneity in PPI networks is akin to differences observed in wiring maps of other complex systems.