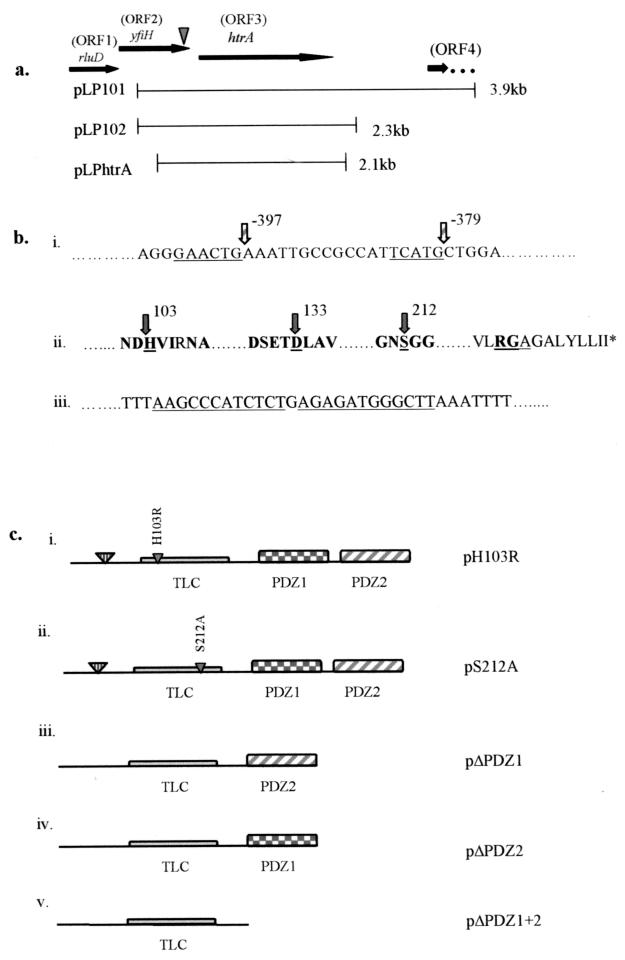
FIG. 3.
Genetic analysis of the disrupted locus in GT20. (a) ORFs and the fragments which were subcloned into pBC to generate plasmids pLP101, pLP102, and pLPHtrA. The inverted arrowhead indicates the location of the kan insert. (b) Conserved sequence elements within promoter regions and predicted HtrA amino acid sequence. (i) Underlined nucleotides indicate the putative ςE −35 and −10 sequences located upstream of the htrA start codon. (ii) Conserved amino acids within the putative catalytic triad as well as conserved surrounding amino acids are indicated in bold. The catalytic triad residues (H, D, and S) are underlined, with arrows and amino acids numbered. The RGA motif is underlined, and the stop codon is indicated by an asterisk. (iii) The perfect direct inverted repeat sequences signifying rho-independent termination downstream of the stop codon are underlined. (c) Conserved domains of HtrA are indicated. The signal sequence cleavage site is indicated by the large inverted triangle, followed by the trypsin-like catalytic domain (TLC) and tandem PDZ domains, designated PDZ1 and PDZ2. Mutant constructs generated are described as follows: (i and ii) Point mutations H103R and S212A, generated by overlap extension PCR to produce the mutant constructs pH103R and pS212A, are indicated by small inverted triangles within the trypsin-like catalytic domain. (iii) Deletion mutant pΔPDZ1, which lacks PDZ domain 1. (iv) Deletion mutant pΔPDZ2, which lacks PDZ domain 2. (v) pΔPDZ1+2, which lacks both PDZ domains.