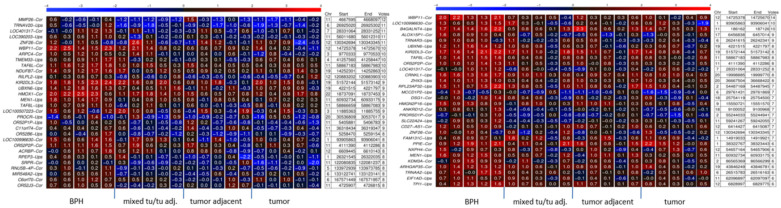
Figure 1.
DNA methylation heat maps of CpG-rich 5′ gene regions from PCa biopsies (G7, 3 + 4) and PCa primary tissue samples (G7, 4 + 3) compared to BPH reference samples. Five tissue samples from BPH prostate glands, five mixed tissue samples of tumor and tumor-adjacent tissue (mixed tu/tu adj.; 50/50%), five samples of tumor-adjacent gland tissue (tumor adjacent) and five tumor tissue samples (tumor, >90%) were used. The left panel corresponds to biopsy samples and the right panel shows the results of primary PCa tissue samples. Only the first 30 most-voted differentially methylated loci out of 237 hypomethylated regions in the biopsies and 202 hypomethylated regions in the primary tumors are presented in each case. Each rectangle in the heat map stands for one CpG-rich 5′ gene region, with its chromosomal location indicated on the right-hand side (Chr, start/end). The “votes” number indicates how many of the samples were found with hypomethylation of the respective region by the democratic method. Redder color corresponds to more methylated regions. I.e., the most unmethylated 5′ gene regions are light blue. The grade of methylation is also indicated by numbers within the colored rectangles. Higher number corresponds to higher methylation. The abbreviation “Cor” after the gene name indicates an examined CpG-rich region of −500 to +500 nucleotides surrounding the transcription start site (TSS). A further examined region of −1500 to −500 bases lying upstream of the TSS is referred to as “Ups” following the gene name.