Figure 1.
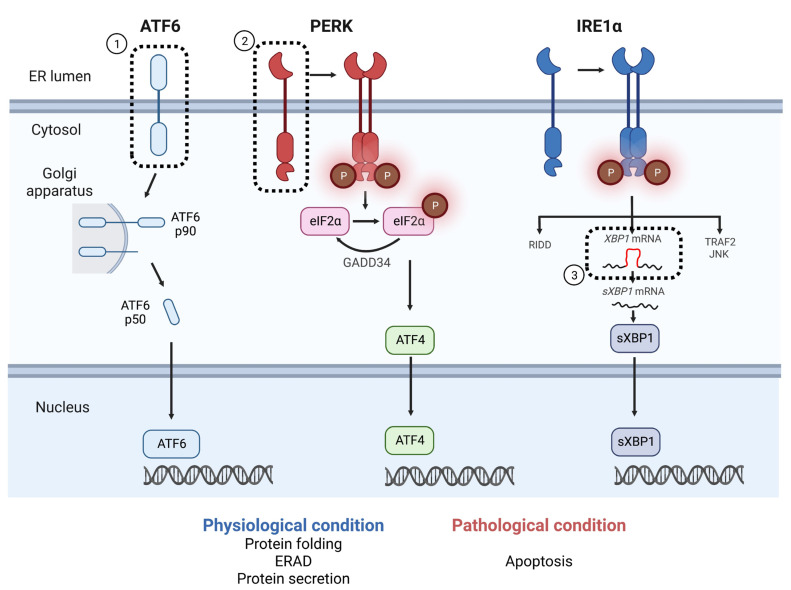
UPR mechanism and the reported SNPs associated with T2DM. The three major regulators (ATF6α, PERK, and IRE1α) are involved in the UPR pathway under pathophysiological conditions. On sensing ER stress, ATF6α (p90) is cleaved at the Golgi apparatus. Cleaved ATF6α (p50) translocates to the nucleus and acts as a transcription factor. PERK arrests protein synthesis by phosphorylating eukaryotic translation initiation factor 2 subunit α (eIF2α). Phosphorylated eIF2α also induces the expression of ATF4, which regulates protein synthesis, apoptosis, and autophagy. IRE1α forms oligomers and autophosphorylates on sensing ER stress, followed by the acquisition of RNase activity. GADD33 dephosphorylates eIF2α and restores protein synthesis. The activated IRE1α splices the XBP1 intron and induces the expression of the transcription factor, spliced XBP1 (sXBP1). IRE1α also suppresses protein folding loads by degrading mRNA, called regulated IRE1-dependent decay (RIDD). In addition, IRE1α activates the TRAF2/JNK pathway followed by the activation of the autophagy pathway. ①–③: The molecules whose SNPs have been reported to contribute to the development of T2DM or insulin resistance include the following. ① ATF6 SNPs, detected in a population of Pima Indians and Dutch Caucasians [12,13]; ② PERK SNPs, detected in Han Chinese populations correlated with increased risk of prediabetes, elevated BMI, and insulin resistance [14]; ③ XBP1 SNPs, reported at high frequency in Han Chinese populations with T2DM and impaired glucose regulation [14,15]—ATF6α (or ATF4), activating transcription factor 6α (or 4); PERK, protein kinase R-like ER kinase; inositol requiring enzyme 1α (IRE1α); ERAD, ER-associated degradation.