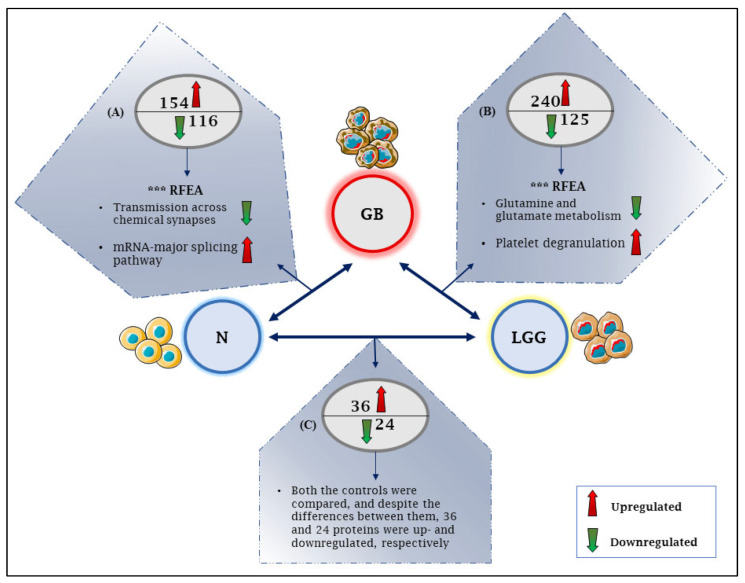
Figure 2.
A schematic representation of differentially expressed proteins in GB as concluded from a meta-analysis across 14 datasets by Tribe et al. GB = glioblastoma, N = normal brain cells, LGG = low-grade gliomas, *** RFEA = most significant parent terms in the reactome functional enrichment analysis. (A) In two or more of the four datasets comparing GB to the normal brain, 154 proteins were upregulated, RFEA of which showed synaptic signaling to be the most significantly over-represented, and 116 proteins were downregulated with mRNA metabolism (mRNA splicing) to be the most significantly over-represented. (B) When comparing GB to LGG, 240 proteins were upregulated, RFEA of which showed platelet degranulation to be overrepresented, and 125 were downregulated in three or more of the ten datasets, with the significant parent term in RFEA involving glutamine and glutamate metabolism. (C) Between LGG and the normal brain controls, 36 proteins were elevated, and 24 proteins were downregulated.