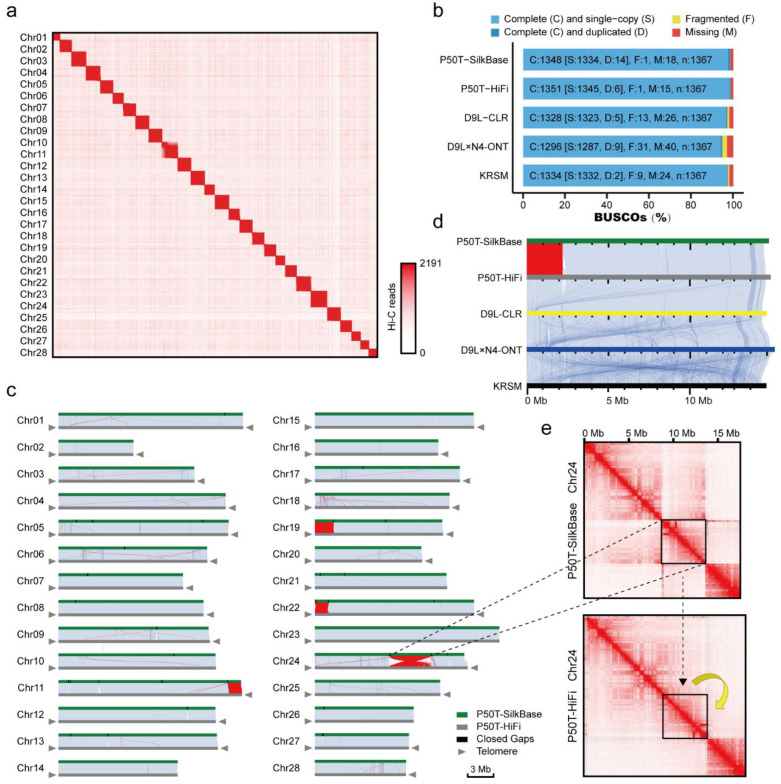
Figure 6.
Summary of different silkworm strains chromosome-level genome assembly. (a) Hi-C genome-wide interaction map of the silkworm (P50T-HiFi) assembly. (b) BUSCO analysis of chromosome-level genome assemblies using the insect odb10 (1367 genes). (c) Collinearity between the silkworm P50T-SilkBase and P50T-HiFi genomes. The synteny blocks are shown by light blue lines. The inversions are indicated by red lines. The telomere sequence repeats are marked by gray triangles. All the P50T-SilkBase gap regions closed in P50T-HiFi are shown as black blocks. The gray triangles indicate the presence of telomere sequence repeats. (d) Collinearity of the P50T-SilkBase, P50T-HiFi, D9L-CLR, D9L × N4-ONT and Korean silkworm (KRSM) Chromosome 19. Synteny blocks are shown by light blue lines. The inversions are indicated by red lines. (e) Hi-C interaction map of silkworm chromosome 24. The black boxed area indicates the error inversion in P50T-SilkBase.