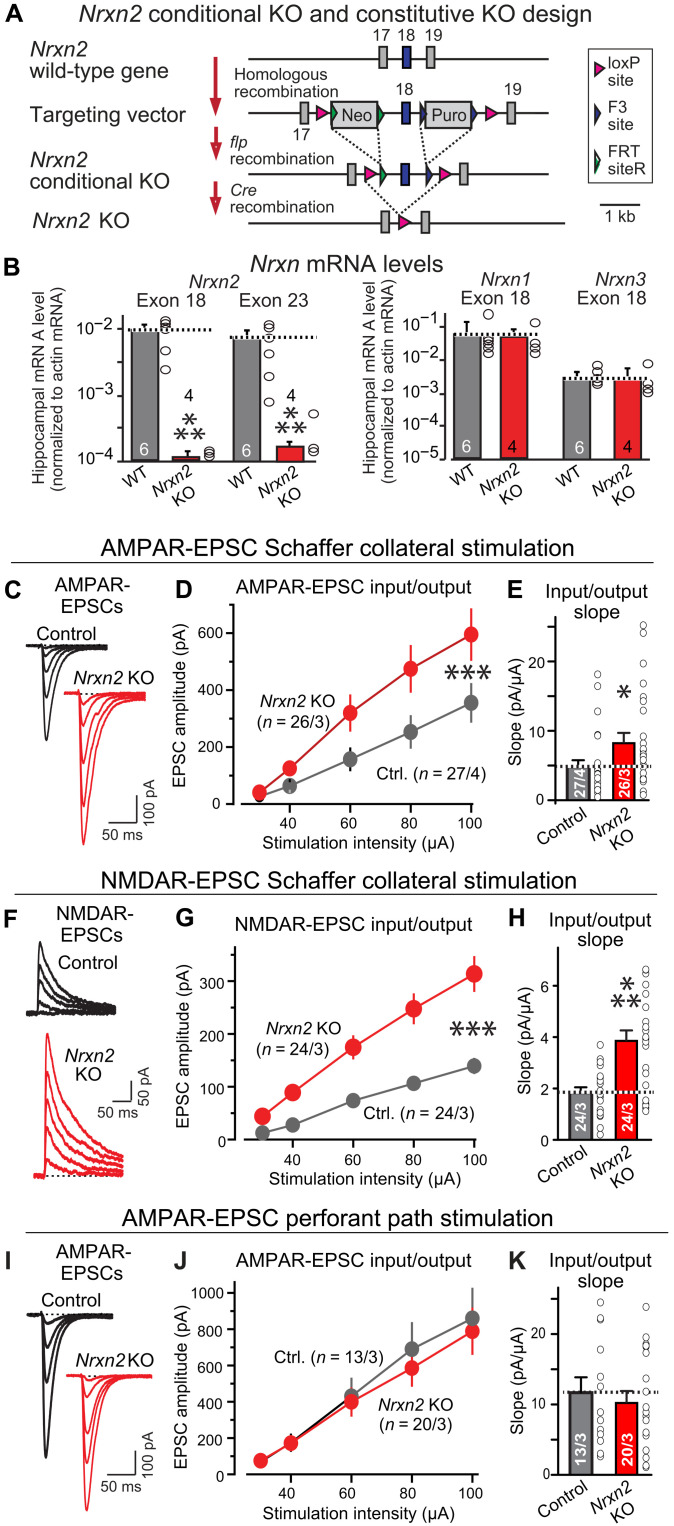
Fig. 1. Constitutive deletion of Nrxn2 increases CA3➔CA1 synaptic connections in the hippocampus.
(A) Nrxn2 cKO and constitutive KO strategy. Two selectable markers [puromycin (Puro) and neomycin (Neo)] were required to obtain embryonic stem cell clones with homologous recombination of the Nrxn2 gene. Exon 18, the first exon shared between Nrxn2α and Nrxn2β, was flanked by loxP sites, enabling Cre-mediated deletion of both Nrxn2α and Nrxn2β. (B) Nrxn2 cKO mice were bred with cytomegalovirus (CMV)–Cre mice to generate littermate wild-type (WT) and constitutive Nrxn2 KO mice. The constitutive Nrxn2 KO suppresses Nrxn2 mRNA levels but leaves Nrxn1 and Nrxn3 mRNA levels unchanged. The exon 18 Nrxn2 mRNA level measurements monitor the exon that is deleted, with the remaining 1% of mRNA detected likely because of background of quantitative reverse transcription polymerase chain reaction (RT-PCR) measurements. The decrease in the exon 23 mRNA levels is likely due to nonsense-mediated decay because the exon 18 deletion should not block Nrxn2 transcription, only the production of a functional protein. (C to E) Nrxn2 KO increases the AMPAR-mediated synaptic responses elicited by Schaffer collateral stimulation [(C) representative traces of AMPAR-EPSCs evoked by increasingly stronger stimuli and recorded at −70 mV in 50 μM picrotoxin and 50 μM D-D-AP-5 (2R)-amino-5-phosphonopentanoate) (2R)-amino-5-phosphonopentanoate); (D) input/output plot of the EPSC amplitude versus stimulus strength; (E) slope of the input/output relation]. (F to H) Nrxn2 KO increases NMDAR-mediated synaptic responses elicited by Schaffer collateral stimulation [same as (C) to (E) except that the responses were recorded at a holding potential of +40 mV in the presence of 20 μM 6-cyano-7-nitroquinoxaline-2,3-dione (CNQX)]. (I to K) Nrxn2 KO has no effect on AMPAR-mediated synaptic responses elicited by stimulation of entorhinal cortex–derived axons [same as (C) to (E)]. Data are means ± SEM; the numbers of neurons per mice analyzed are listed in bar graphs. Statistical assessments were performed by the Mann-Whitney test comparing KO to control (B, E, H, and K) or by two-way analysis of variance (ANOVA) tests (D, G, and J), with *P < 0.05, **P < 0.01, and ***P < 0.001. Ctrl., control.