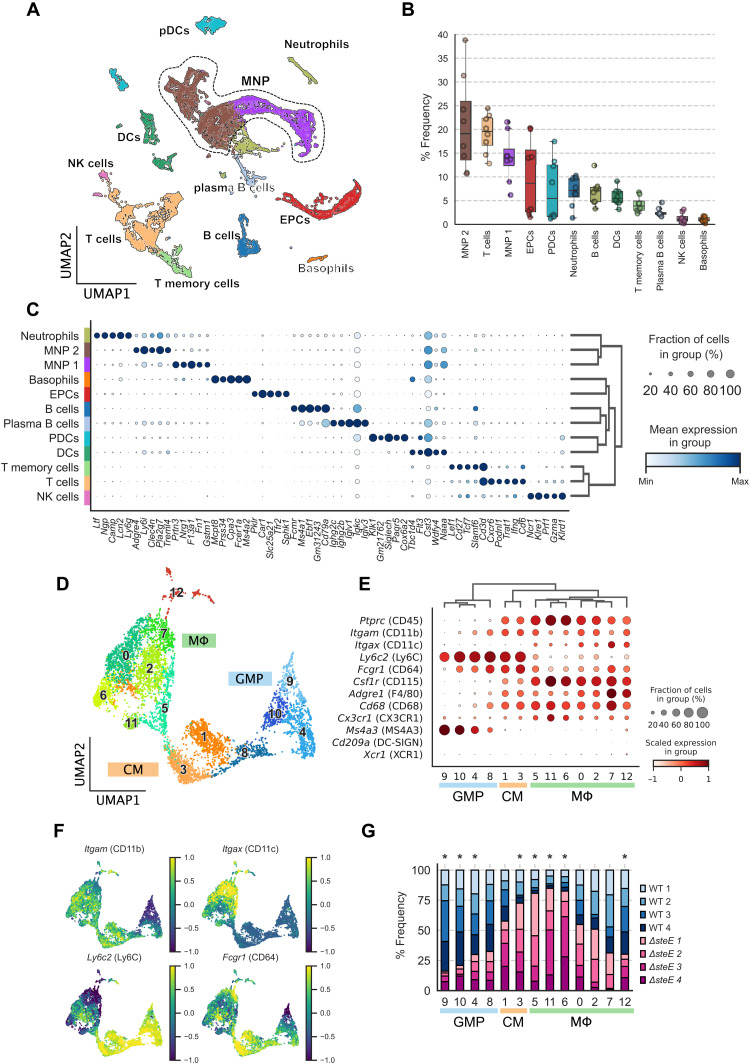
Fig. 1. The spectrum of splenic macrophages, monocytes, and their precursors during persistent STm infection.
Splenocytes from mice infected with either WT STm or ΔsteE STm for 4 weeks were permissively FACS-sorted to simultaneously enrich for MΦs and capture all immune cell types for scRNA-seq. (A) Uniform Manifold Approximation and Projection (UMAP) of sequenced cells from both WT STm- and ΔsteE STm-infected spleens. DCs, dendritic cells; pDCs, plasmacytoid DCs; NK cells, natural killer cells; EPCs, erythroid precursor cells. (B) Percent frequencies for each immune cell type. (C) Top five DEGs in each immune cell population. Radius and color intensity of the dots reflect detection rate and mean expression for each gene, respectively. (D) UMAP projection of MNPs, excluding dendritic cells and neutrophils, colored by the assigned subpopulations: GMPs, CMs, and MΦs. (E) Dotplot showing expression levels of myeloid marker genes on GMP, CM, or MΦ clusters. (F) Expression levels of Itgam (CD11b), Itgax (CD11c), Ly6c2 (Ly6C), and Fcgr1 (CD64) previously shown to express on MΦs that densely populated STm granulomas (fig. S1B). (G) Differential representation test for GMP/CM/MΦ clusters in WT STm- and ΔsteE STm-infected animals. Asterisk above the bar indicates a greater than twofold difference in representation ratio and statistical significance in association with bacterial strain based on differential representation test (FDR < 0.05).