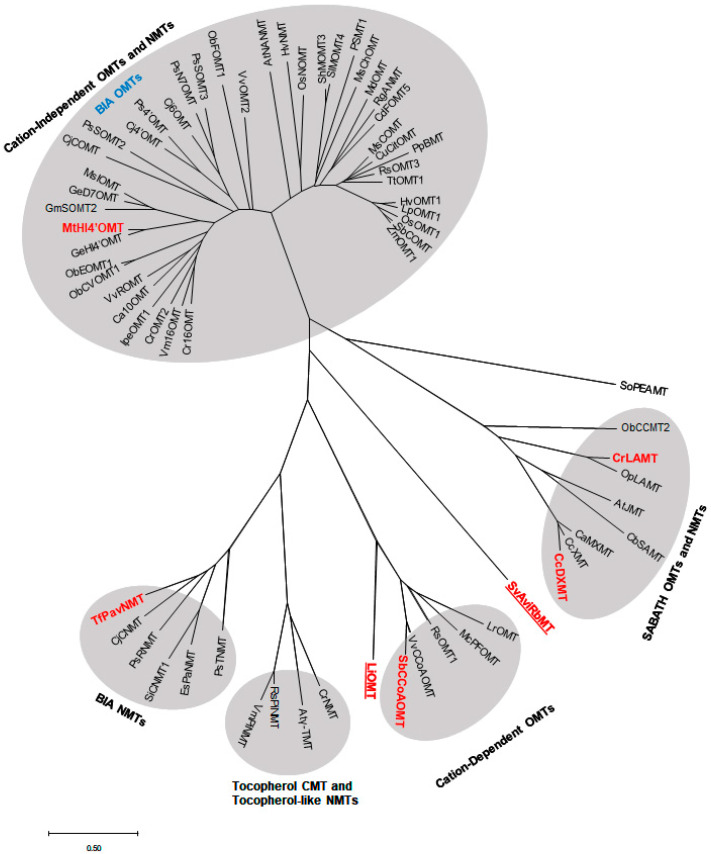
Figure 4.
Phylogenetic relationships among functionally characterized plant NPMTs and bacterial MTs. A phylogenetic tree was generated using the neighbor-joining method and the Poisson correlation method via MEGA X [161]. The scale bar shows the amino acids substituted per sequence alignment. Bolded sequences indicate the NPMTs further analyzed in this review. The underlined sequences are bacterial methyltransferases, showing the relationship between bacteria and plant NPMTs. The plant species and accession numbers can be found in Table 1. The bacterial MT species and accession numbers are: LiOMT, Leptospira interrogans O-methyltransferase (WP_000087781); SvAviRbMT, Streptomyces viridochromogenes antibiotic-resistant mediating O-methyltransferase (WP_003998250).