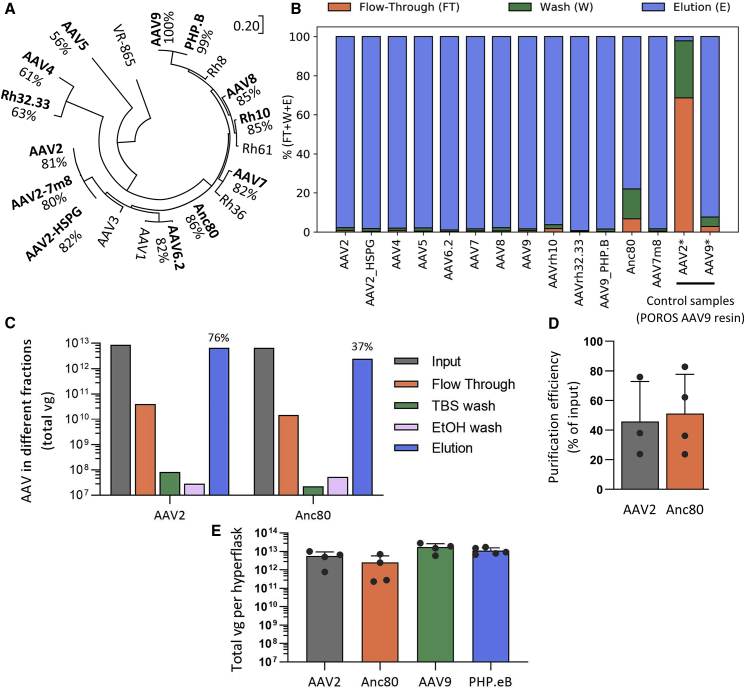
Figure 1.
AAV purification using AAVX affinity chromatography
(A) Phylogeny depicting the diversity of AAV capsids included in this report (bold) along with the percent identity (by amino acid) compared with AAV9. The tree is drawn to scale with branch lengths depicting substitutions per site. VR-865 is an avian AAV used as an outgroup. (B) Affinity of AAVX to various AAV serotypes tested in a static binding assay. The flow-through (FT), wash (W), and eluted fractions (E) were collected and analyzed by qPCR to quantify their vector genome copies. Data represented as percent vector genomes (vg) of the input. Each serotype was applied to unused AAVX resin. (C) AAV purification of AAV2 and Anc80 using AAVX resin in an HPLC setting. Fractions were taken from input, flow-through, at Tris-buffered saline (TBS) and ethanol wash steps and at elution, and AAV content was quantified using qPCR. Percent recovery for these purifications is shown above elution bars. N = 1 each. (D) Average purification efficiencies of AAV2 and Anc80 (percent recovery of AAV in the elution). (E) Total yields of purified AAV2, Anc80, AAV9, and PHP.eB preps with no optimization of the process. Each dot represents an AAV prep from one HYPERFlask (1,720 cm2 growth area). Error bars denote standard deviation. All purifications were carried out at room temperature, using 1-mL AAVX column at 1 mL/min flow rate. All values estimated are above qPCR limit of detection (approximately 105 vg/mL).