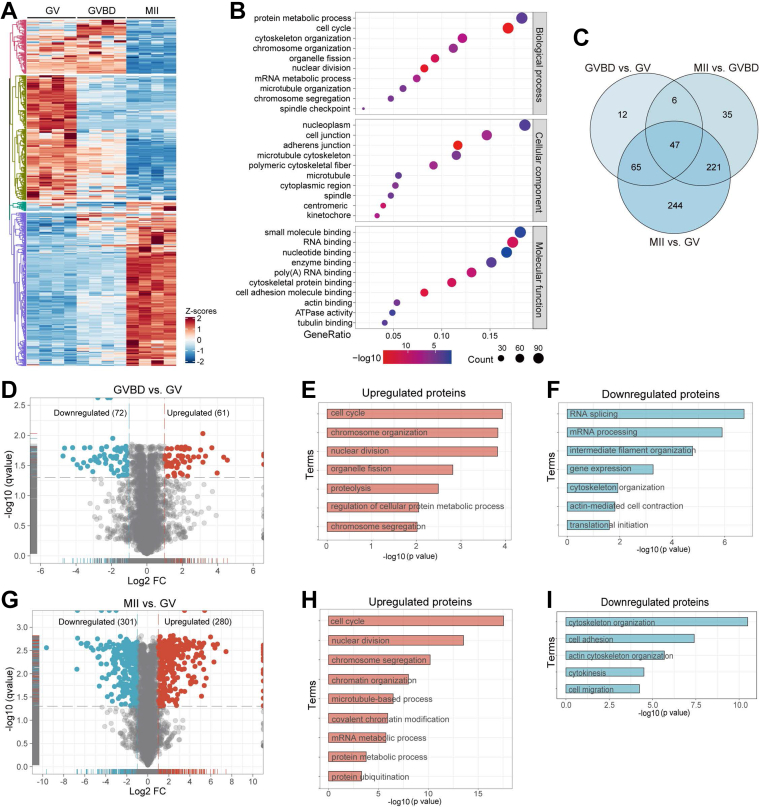
Fig. 2.
Functional Analysis of Differentially Expressed Proteins.A, heatmap showing the expression profile of 634 differentially expressed proteins (DEPs, fold change>2, false discovery rate <0.05) during oocyte maturation. B, gene ontology (GO) enrichment analysis of DEPs was performed using the Pathway Studio (V6.00) software. C, Venn diagram showing the exclusive or shared DEPs between three different stages. D, volcano plot showing DEPs (downregulated, blue; upregulated, red) in GV and GVBD oocytes. (E-F) GO enrichment analysis of upregulated and downregulated DEPs in GV and GVBD oocytes. G, volcano plot showing DEPs (downregulated, blue; upregulated, red) in GV and MII oocytes. H–I, GO enrichment analysis of upregulated and downregulated DEPs in GV and MII oocytes.