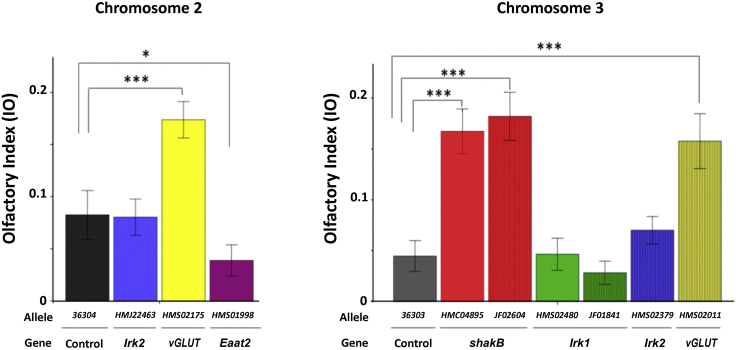
Figure 5.
Effects of RNAi silencing of several candidate genes in MZ317 cells on olfactory behavior in response to ethyl acetate (10−1.5)
RNAi lines were generated by the TRiP program, which also produced control lines for chromosome 2 and chromosome 3. We used 2 UAS-RNAi stocks for each gene except for Eaat2. In all cases, we compared the experimental flies, Mz317-Gal4/UAS-RNAi-GeneX, against the corresponding controls, Mz317-Gal4/UAS-36304 and MZ317-Gal4/UAS-36303, for chromosomes 2 and 3, respectively. The mean ± SEM values are presented. N = 20 for each fly type. T test values are from left to right; for chromosome 2, t-Irk238 = 0.107, n.s.; t-Eaat238 = 2.226∗; t-vGLUT38 = −4.417∗∗∗; for chromosome 3, t-shakB-HM38 = −6.491∗∗∗; t-shakB-JF38 = −6.912∗∗∗; t-vGLUT38 = −5.159∗∗∗; t-Irk1-HM38 = −0.194, n.s.; t-Irk1-JF38 = −1.122, n.s.; t-Irk2 38 = −1.172, n.s. Significant differences of each fly type compared with the control flies are indicated in the figure (∗∗∗ = p < 0.001; ∗∗ = p < 0.01; ∗ = p < 0.05).