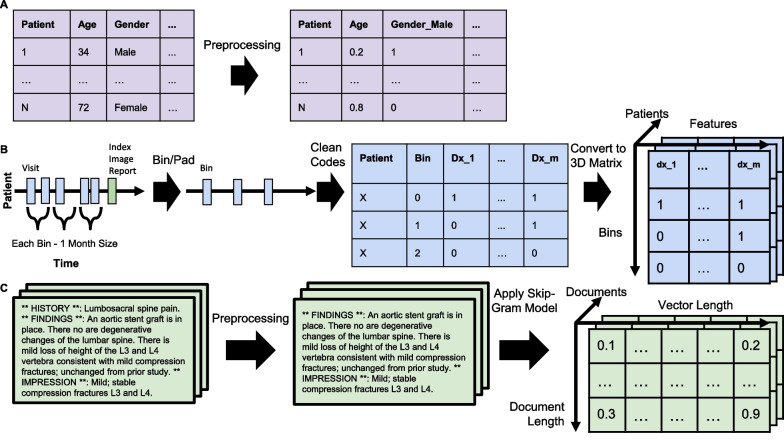
Fig. 2.
Visualization of data preprocessing for deep learning. A For the demographics data (i.e. static data), we created dummy variables for the categorical features and normalized the discrete numerical feature (i.e. age) at the patient level. B For the diagnosis, procedures, and drug names data for the deep learning model (i.e. temporal data), we limited the information to the last three months of information prior to the index image for both prediction tasks. We cleaned up the ICD codes by mapping them to level three in the hierarchy. To maintain the same number of bins (i.e. three), we added empty bins to patients with less than three bins. Finally, we converted the dataframe into a 3D tensor. C We pre-trained a skip-gram model on 123,461 LIRE reports. We applied our model to each index imaging report to extract a feature representation. ICD, International Classification of Diseases; LIRE, Lumbar Imaging with Reporting of Epidemiology