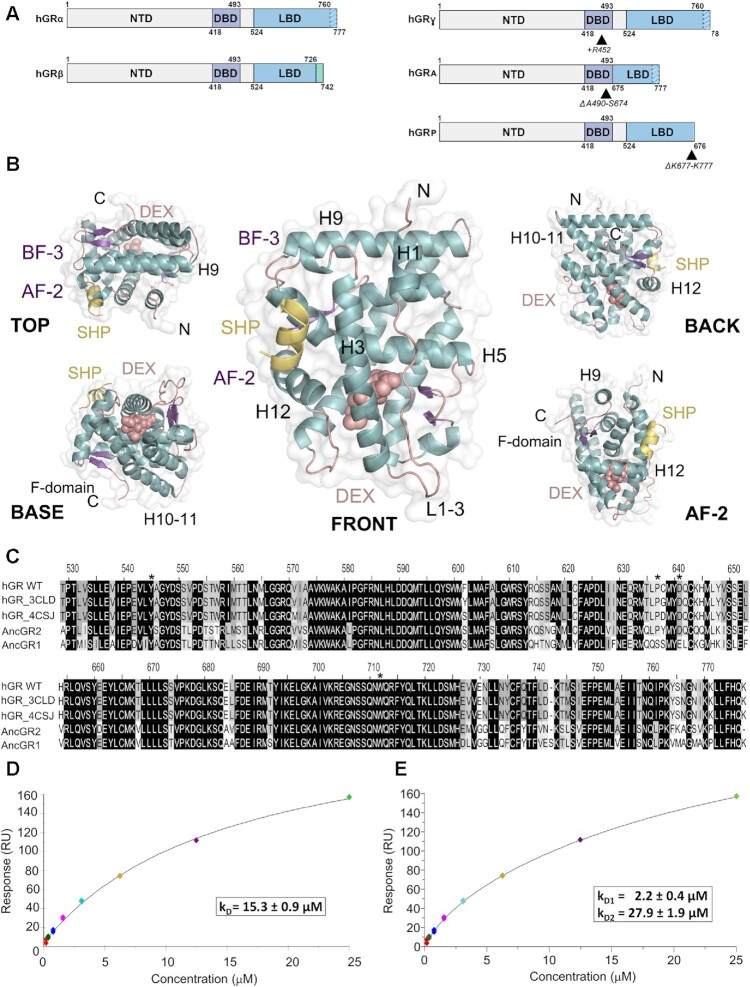
Figure 1.
The ligand-binding domain of GR self-associates in solution. (A) Schematic representation of GR domain organization. GRα and GRβ are identical up to residue 727 (H10/11), but the last 50 (in GRα) and 15 residues (in GRβ, green box) are unrelated. Other GR isoforms are shown (right). (B) Overall structure of the GR-LBD monomer. The module is shown in standard orientation in the middle of the panel (i.e. with H1 and H3 displayed facing the viewer and the AF-2 pocket on the left). Four additional views are shown to present other LBD areas. Models are depicted as cartoons with helices (blue), loops (pink) and beta-sheets (purple). DEX (salmon spheres) and SHP peptide (yellow cartoon) are shown. The BF-3 pocket is also labeled. (C) Sequence alignment of LBDs between WT human GR, two engineered variants used in several structures (PDBs 3CLD and 4CSJ), and the ancGR1 and 2 forms. Strictly conserved residues are white with black shading; other conservatively replaced residues are shaded gray. Residues mutated in the current study are marked with asterisks. (D, E) SPR analysis of GR-LBD self-association according to (D) 1:1 or (E) multisite models. The results of experiments conducted in duplicate are shown along with the calculated affinity constants (kD). Data were fitted to the 1:1 and multisite models with χ2 values of 4.52 and 1.25, respectively.