Figure 3.
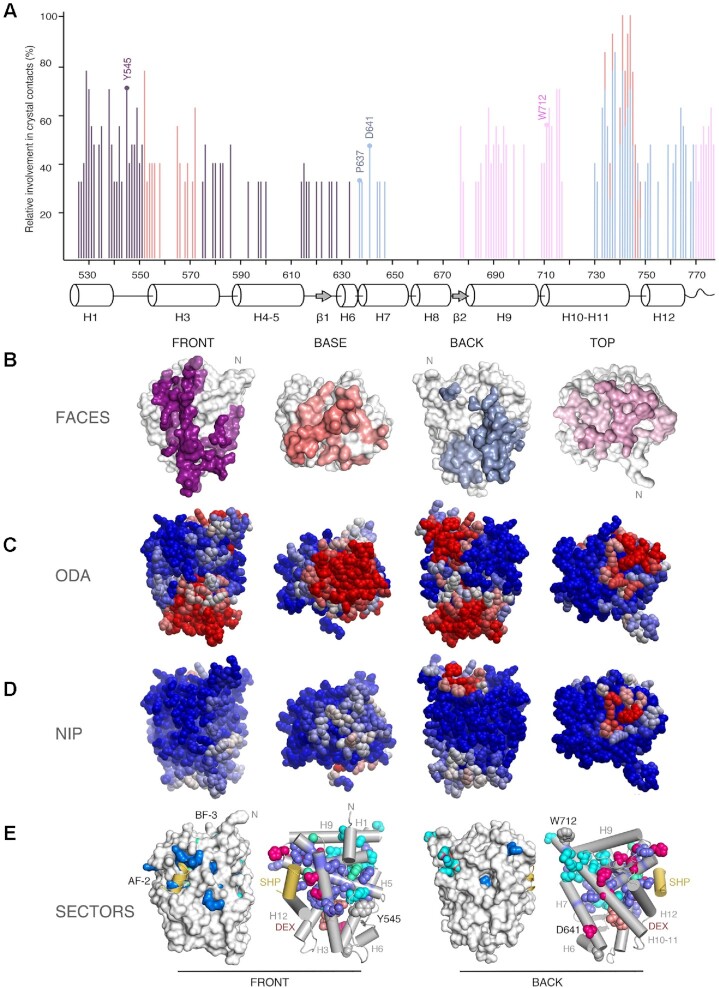
Experimental structures and bioinformatics analyses unveil four major homodimerization surfaces on GR-LBD. (A) Relative frequencies of residue involvement in GR-LBD homodimer formation. Bar height indicates how often a given residue engages in crystal contacts in all available structures of GR-LBD, normalized to the residue most frequently found in homodimer interfaces, Leu741. Secondary structure elements given below the plot correspond to the crystal structure of human GR-LBD resolved at the highest resolution, 6NWL. (B) Residues involved in GR-LBD homodimerization cluster in continuous patches on its front (colored purple), base (coral), back (blue) and top (pink) faces. The association of these four faces yields the catalog of GR-LBD dimers represented in Figure 4. Models are shown in the same orientation and at the same magnification in panels (C)–(E). (C) Predicted protein-protein interaction ODA. ODA ‘hotspots’ (residues with favorable docking energy; ODA < –10.0 kcal/mol) are colored red, residues with ODA > 0 kcal/mol are shown in blue, and intermediate values are scaled accordingly. ODA hotspots form continuous surface patches that essentially overlap with the four protein-protein interaction interfaces shown in panel (B). (D) Hotspot interface residues predicted from docking experiments. Surface residues are colored according to their NIP. Residues with NIP > 0.4 and < 0 are colored red and blue, respectively; intermediate values are scaled accordingly. (E) SCA identifies two sectors of clustered, physically connected residues in GR-LBD. The front and back orientations of GR-LBD are depicted, and in both cases the module is represented as a solid surface and as a cartoon, with helices shown as rods and labeled. Residues belonging to sectors I and II are shown with their side chains atoms as spheres, colored cyan and dark blue, respectively. Residues belonging to both sectors are colored pink. SHP peptide is colored yellow, and DEX is shown as salmon spheres.