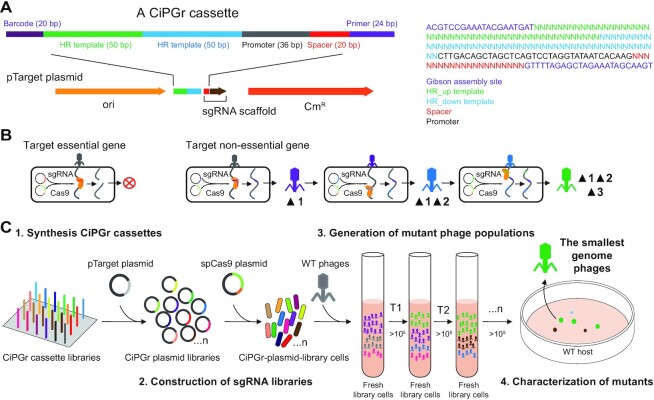
Figure 1.
Schematic diagram illustrating the CiPGr approach. (A) Design of the CiPGr cassette. Heterogeneous barcodes (20 bp) were designed and used to separate the cassettes into eight libraries by PCR using primer (24 bp) sequences; the primer was part of the sgRNA sequence and was designed as the Gibson assembly site for the cassettes and the pTarget plasmid. CmR, chloramphenicol resistance gene; HR, homologous recombination; ori, origin of replication. (B) Continuous genome editing of the phages using the dual-plasmid system. Essential genes were resistant to editing; non-essential genes were deleted or disrupted, and mutations accumulated in the phage genome. (C) Workflow of the CiPGr approach. Step 1: the CiPGr cassettes were synthesized on a DNA chip. Step 2: the CiPGr cassettes were inserted into pTarget and delivered to MG1655 or Salmonella (ST56) host strains containing the spCas9 plasmid, resulting in eight CiPGr plasmid libraries. Step 3: the iterative CiPGr process was initiated by adding WT phages (T7, seszw, T4 and selz) to cultured cells carrying the libraries, followed by repeated transfer of the mutant phage populations to fresh cultures. Step 4: the mutant phages were isolated, characterized and sequenced, and those with the smallest genomes were identified.