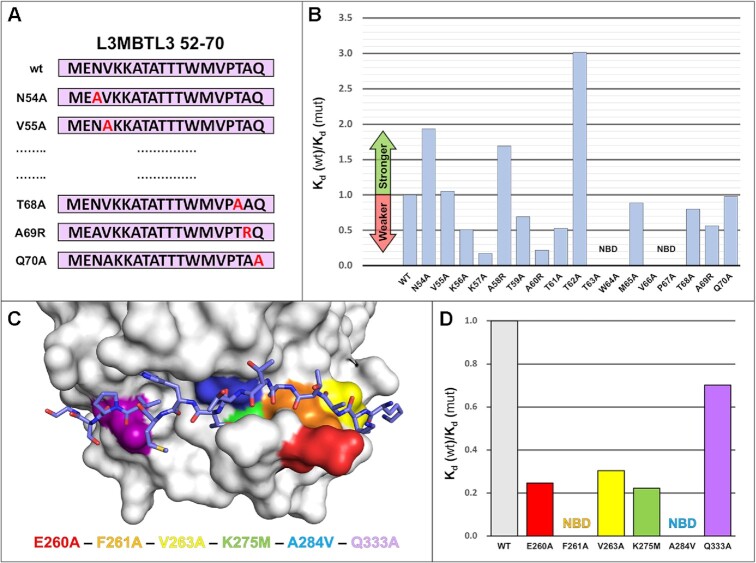
Figure 5.
RBPJ–L3MBTL3 binding analysis reveals residues sensitive to mutation. (A) Diagram of scanning point mutants of the 19-mer L3MBTL3 RBP-ID (52–70) for ITC binding studies. Starting at N54, each residue was mutated individually to an alanine, while native alanines were mutated to arginine. (B) Affinity change plot [Kd (wt)/Kd (mut)] for L3MBTL3 alanine scanning mutants tested against RBPJ wild-type (WT). Changes are plotted as the ratio of Kd values, where an increase in Kd (weaker binding) is below 1 and vice versa. Mutations along the length of the peptide have varying effects on binding, with K57A and A260R significantly affecting binding; T63A, W64A, V66A, from the hydrophobic tetrapeptide region, and the adjacent P67A mutant all completely abrogate binding. N54A, A58R, and T62A show increased binding to RBPJ, with T62A from the dithreonine loop behaving as the tightest binding peptide (∼300 nM Kd). (C) Visualization of BTD residues targeted for mutation with color coding. The L3MBTL3 peptide is colored purple and shown as sticks. These BTD residues (E260, F261, V263, K275, A284, Q333) have all been shown to influence binding of other RAM-like coregulators, with F261 and A284 mutations causing the largest decreases in affinity. (D) Affinity change plot for BTD mutants tested against L3MBTL3 52–70 wild-type peptide. All mutations negatively affect binding, with Q333A having the smallest effect; F261A and A284V completely abrogate L3MBTL3 binding; and E260A, V263A and K275M have modest effects on binding. NBD = no binding detected.