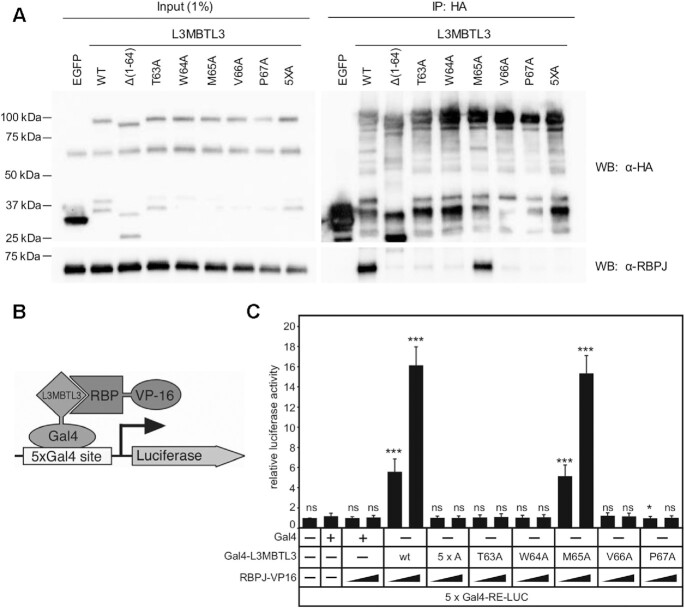
Figure 6.
Cellular analysis of L3MBTL3 mutants. (A) Figure shows western blot (WB) of immunoprecipitated (IP) HA-tagged L3MBTL3, wild-type (WT) and mutants, with RBPJ from U87-MG glioblastoma cells. The N-terminal deletion construct L3MBTL3 (Δ1–64) has previously been shown to completely abrogate interactions with RBPJ in cells (22). The binding deficient L3MBTL3 point mutants identified by ITC (T63A, W64A, V66A, P67A) are similarly impaired for binding RBPJ in cells; whereas, L3MBTL3 M65A, which retains 90% of binding in vitro, only modestly affects interactions with RBPJ in cells. As expected, the 5× alanine mutant (5XA = T63A/W64A/M65A/V66A/P67A) completely abrogates L3MBTL3–RBPJ interactions. (B) Schematic representation of mammalian two-hybrid assay. HeLa cells were cotransfected with Gal4-L3MBTL3 wild-type and mutant constructs and increasing amounts of RBP-VP16 together with pFR-Luc containing Gal4 recognition sites (5xGal4-RE-LUC). Luciferase activity was determined from 100 μg portions of total cell extract. Fold-activation was determined by the relative luciferase activity after cotransfection of the Gal4 construct alone. Mean values and standard deviations from four experiments are shown. (C) Gal4-L3MBTL3 and RBPJ-VP16 interact to induce luciferase expression in an RBPJ-VP16 concentration dependent manner. Luciferase expression in the mutants corroborates the binding data and coimmunoprecipitation results, with only the M65A mutant able to recruit RBPJ-V16 to activate the reporter.