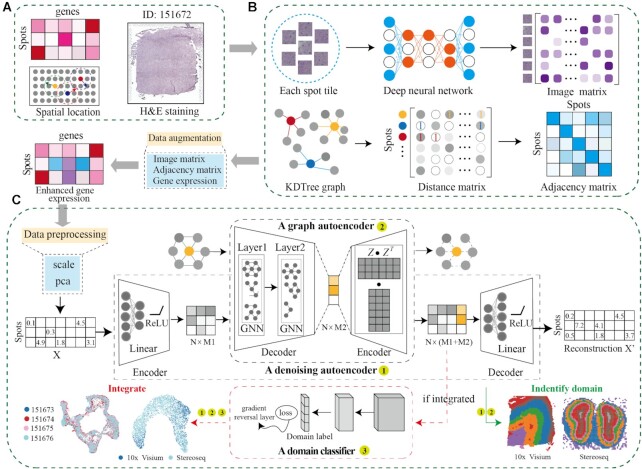
Figure 1.
Workflow of the DeepST algorithm. (A) DeepST workflow begins with ST data, taking hematoxylin and eosin (H&E) staining (optional), spatial coordinates, and spatial gene expression as input. (B) DeepST initially uses the H&E staining to collect tissue morphological information, then normalizes the gene expression of each spot based on similarity against adjacent spots using a pre-trained deep learning model. Morphological similarity between adjacent spots is calculated by this matrix, and the weights of gene expression and spatial location are merged to re-assign an augmented expression value for each gene inside a spot. (C) DeepST generates three network frameworks, where a denoising autoencoder network and a variational graph autoencoder are used to extract the final latent embeddings, and a domain discriminator is used to fuse spatial data from various distributions (red dotted box, the part only for integration tasks).