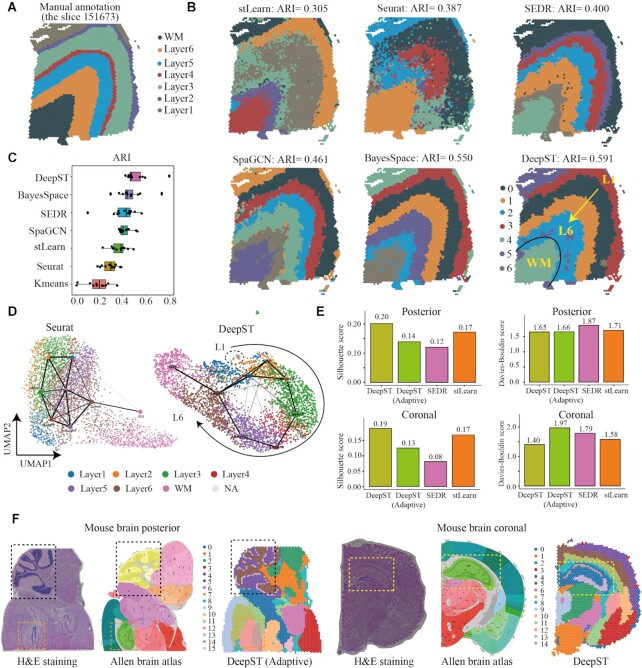
Figure 2.
DeepST improves spatial domain recognition in brain tissue. (A) DLPFC layers were annotated by Maynard et al.(23). The ground truth of spots was mapped on their spatial position in slide 151673, which was separated into six cortical layers (L1–L6) and white matter (WM). Layers with annotations are provided on the remaining slides (Supplementary Figures 2–12). (B) Identification of spatial domains by DeepST, and existing state-of-the-art algorithms (BayesSpace, SpaGCN, SEDR, stLearn, Seurat and K-means) algorithms for slide 151673. (C) Boxplot of the performance of DeepST and other algorithms for all 12 DLPFCs. The x-axis shows the adjusted rand index (ARI), which was used to compare the similarity of the predicted spatial layers and the manually annotated layers for each algorithm. (D) UMAP visualizations and PAGA graphs were generated for slide 151673 with Seurat-derived principal components (left) and DeepST-derived embeddings (right). (E) Histograms of Silhouette Coefficient (SC) and Davies-Bouldin (DB) scores for mouse brain posterior and coronal data, respectively, including algorithms DeepST, DeepST (Adaptive, means no prior knowledge), SEDR and stLearn. (F) Spatial domains of mouse brain tissue sagittal posterior and coronal regions. The H&E staining generated from raw data (left); The corresponding anatomical Allen Mouse Brain Atlas (middle, https://atlas.brain-map.org/); Spatial domains identified by DeepST (right). The yellow box denotes the cornu ammonis and dentate gyrus areas in the coronal portion; The black box denotes the cerebellar cortex; The orange box denotes dentate gyrus areas in the sagittal posterior.