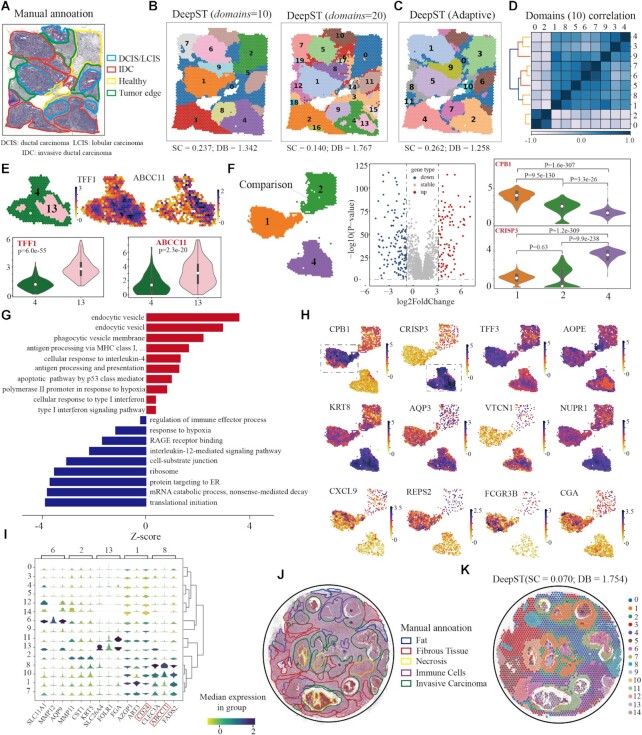
Figure 4.
DeepST can dissect spatial domains from cancer tissue at a finer level. (A) Visium spatial transcriptomics data of a breast cancer sample annotated by pathologists. IDC, invasive ductal carcinoma; DCIS, ductal carcinoma in situ; LCIS, lobular carcinoma in situ; tumor edge; healthy region. (B) Spatial domains identified by DeepST on human breast cancer (Block A, Section 1) with domains= 10 and domains= 20. (C) Spatial domains were identified without a priori knowledge by DeepST. (D) Heatmap of Pearson correlation coefficient between domains (domains= 10). (E) Expression of TFF1 and ABCC11 with regional annotation on left (top); Violin plots of the two genes(bottom). (F) Differential expression analysis among domains 1, 2 and 4. Spatial location of domains 1, 2 and 4 (left); Volcano graph of DEGs between domains 1 and 4 (domains= 10) (middle); CPB1 and CRISP3 express differentially between domains 1 and 4 (purple) (right). (G) Gene ontology enrichment analysis of the DEGs between domains 1 and 4. Red denotes pathway with upregulated genes in domain 1; blue is the opposite. (H) Visualizations of the DEGs (|log fold change| 2) between domains 1 and 4 with k= 10. T-test on the means of two independent domains. (I) Stacked violin plot show expression of all domains on the top three DEGs of domains 6, 2, 13, 1 and 8. (J) H&E of human breast cancer sample annotated by Agoko's telepathology platform. (K) Visium spatial transcriptomics with spatial domains generated by DeepST.
2) between domains 1 and 4 with k= 10. T-test on the means of two independent domains. (I) Stacked violin plot show expression of all domains on the top three DEGs of domains 6, 2, 13, 1 and 8. (J) H&E of human breast cancer sample annotated by Agoko's telepathology platform. (K) Visium spatial transcriptomics with spatial domains generated by DeepST.