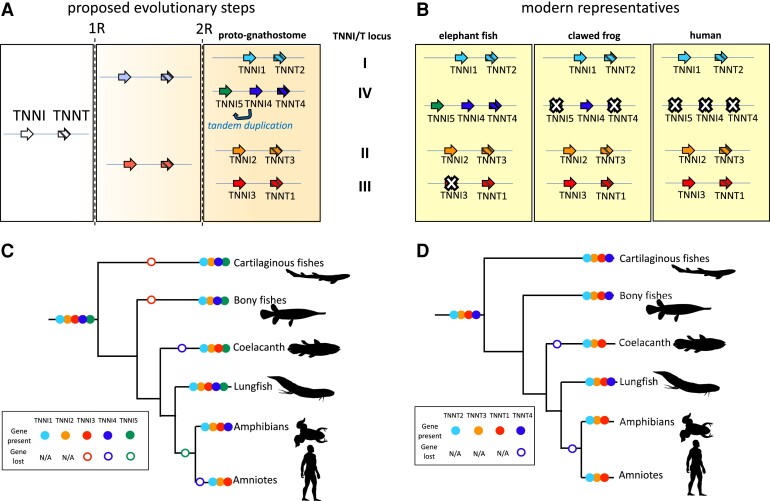
Fig. 8.
The origin and losses of TNNI and TNNT genes in vertebrates. (A) Predicted evolutionary steps in evolution showing how whole-genome duplications (1R and 2R) and tandem duplications generated gene diversity. White crosses (X) mark genes that have been lost in extant lineages. Whilst our maximum-likelihood tree suggests the tandem duplication giving rise to TNNI4 and TNNI5 occurred before the genome duplication that gave rise to TNNI1 and TNNI5, which would require subsequent gene loss of a paralog in both gnathostomes and cyclostomes independently, it is also possible that the tandem duplication occurred only in TNNI4 and TNNI5 after 2R (here shown by arrow marked “tandem duplication’). (B) Comparison of which duplicated genes were retained or lost in three specific gnathostomes (elephant shark, Ca. milii, tropical clawed frog, X. tropicalis and human, H. sapiens. (C and D) broader overview of patterns of TNNI and TNNT gene loss in major vertebrate lineages. Animal silhouettes are courtesy of phylopic.org.