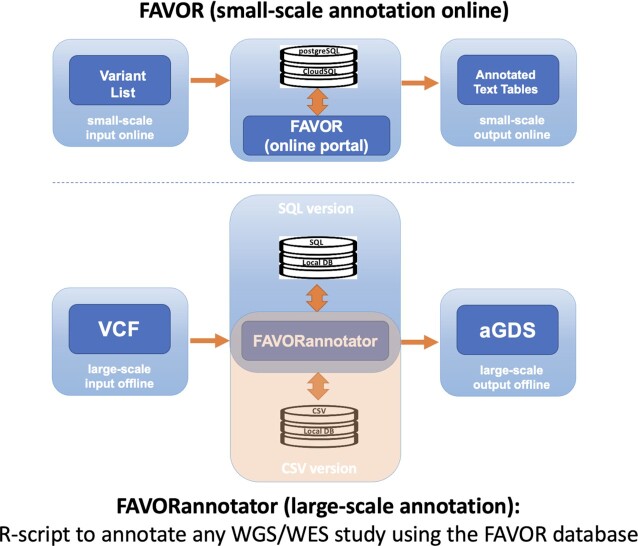
Figure 7.
Graphical representation of the features of FAVOR batch annotation and FAVORannotator. For small-scale annotation (up to 10 000 variants), batch annotation can be used for online annotation at the FAVOR website. For large-scale annotation, e.g. hundreds of millions of variants in a Whole Genome/Exome Sequencing (WGS/WES) study, FAVORannotator can be used for annotation in a local cluster or a cloud platform, e.g. Amazon Web Services (AWS) and Google Cloud Platform (GCP). FAVORannotator uses the FAVOR backend database, which is available in the SQL or CSV formats, and outputs an aGDS file that integrates genotype and annotation data in a single file.