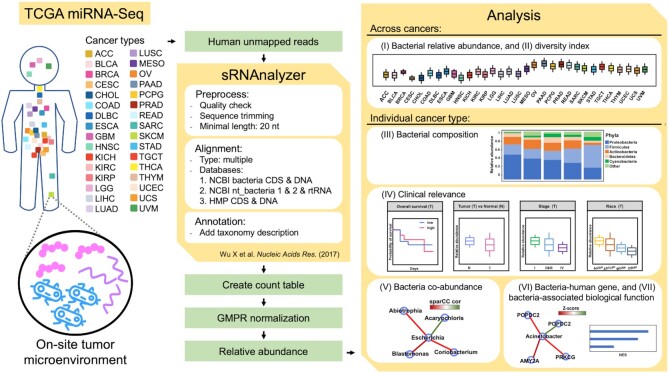
Figure 1.
An overview of BIC analysis workflow. We downloaded miRNA-seq data from TCGA and used sRNAnalyzer for read processing. We parsed and merged count tables, conducted GMPR normalization, and produced the bacterial relative abundance matrixes of each taxonomic level in our scripts. We provide seven modules in the BIC Analyses panel for users to query and download results.