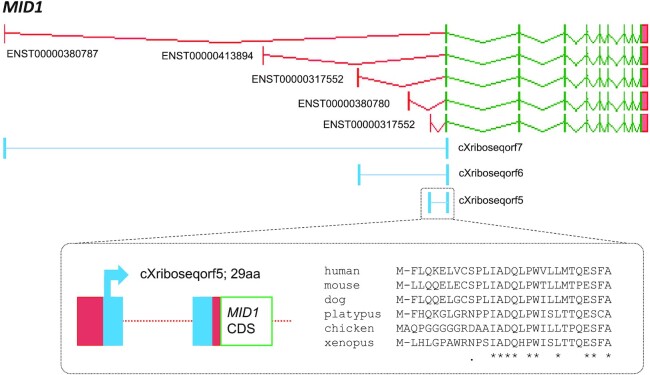
Figure 1.
Upstream open reading frames in MID1. GENCODE 41 annotation includes five distinct transcript start site regions within midline 1 (MID1), a TRIM-family protein-coding gene. Five representative transcripts are shown here; additional transcriptional complexity has been omitted for clarity. Three replicated Ribo-seq ORFs are located on three transcripts: cXriboseqorf7, cXriboseqorf6 and cXriboseqorf5 found respectively on ENST00000380787, ENST00000317552 (the MANE Select transcript) and ENST00000317552. Each is a translated uORF, with three distinct first exon ORF portions being linked to a shared second exon ORF portion. The shared ORF portion has a positive PhyloCSF score, indicating that it has evolved as a protein-coding sequence. However, PhyloCSF only supports the protein-coding potential of one of the three alternative first exons, cXriboseqorf5 on ENST00000317552. A multispecies protein alignment (inset) finds that cXriboseqorf5 has intact orthologs across tetrapods with accompanying transcript support; beyond the five representative species shown, the ORF appears potentially conserved across vertebrates. This ORF has thus been annotated as the new protein-coding gene ENSG00000291314. In contrast, the first exon ORF portions of cXriboseqorf7 and cXriboseqorf6 present equivocal evolutionary signatures, lacking PhyloCSF support to indicate protein-level function. Nonetheless, cXriboseqorf7 at least is conserved as an ORF in mammals as well as reptiles and avians, and if this ORF is not protein-coding it may turn out to have a regulatory function that is evolving under a different mode of selection. This may also be true of cXriboseqorf6, and in fact, we do not rule out the possibility that both cXriboseqorf7 and cXriboseqorf6 encode functional proteins in spite of the lack of PhyloCSF support.