Figure 1.
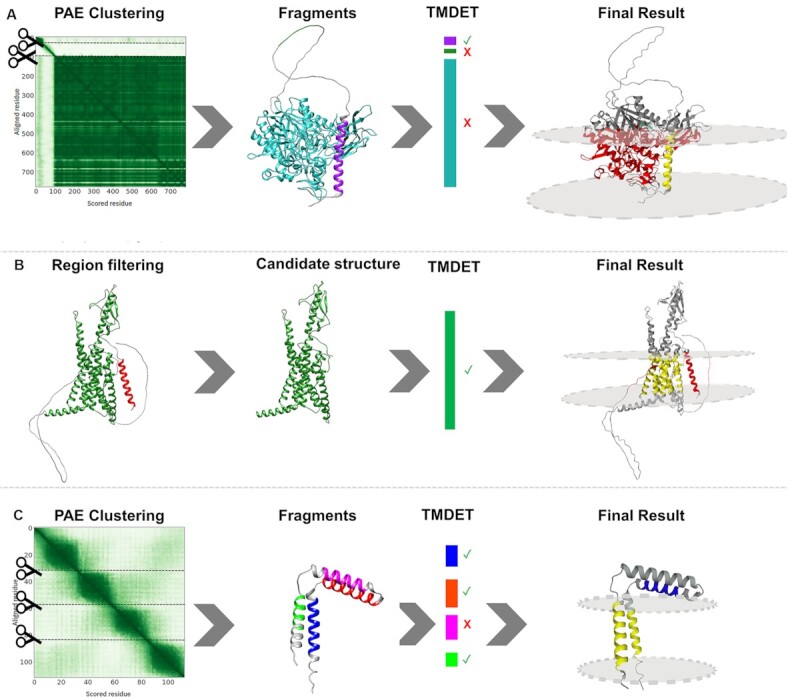
(A) ASAH_HUMAN (Q9NR71) Left to right: The structure was sliced into potential domains based on the Predicted Alignment Error matrix; The structure fell apart into three fragments: 2 domains (purple and cyan) and a flexible linker (green); TMDET detected the membrane plane in the helical domain (purple); The final result shows the TM helix (yellow), however, the other globular fragment is not compatible with the membrane plane (red). (B) PTH2R_HUMAN (P49190) Left to right: signal peptide (red) and low reliability regions (grey) are detected; These regions might penetrate the membrane, therefore they are masked out; TMDET detects the membrane plane in the structure; The final result shows the TM domain (yellow), while masked out segments embedded in the membrane are highlighted (red). (C) TM14C_HUMAN (Q9P0S9) Left to right: The structure was sliced into potential domains based on the Predicted Alignment Error matrix; The structure fell apart into four helical fragments (blue, orange, purple and green); TMDET detected membrane helices in three fragments; The membrane plane cannot be positioned in a way that all three helices are included. Green and blue fragments are in the membrane plane (yellow), the extra TM segment is highlighted (blue). Notably, in comparison with the experimental structure (2los_A), the orientation of the N-terminal TM helix is wrong, one TM segment is erroneously predicted to lie parallelly to the membrane plane.
