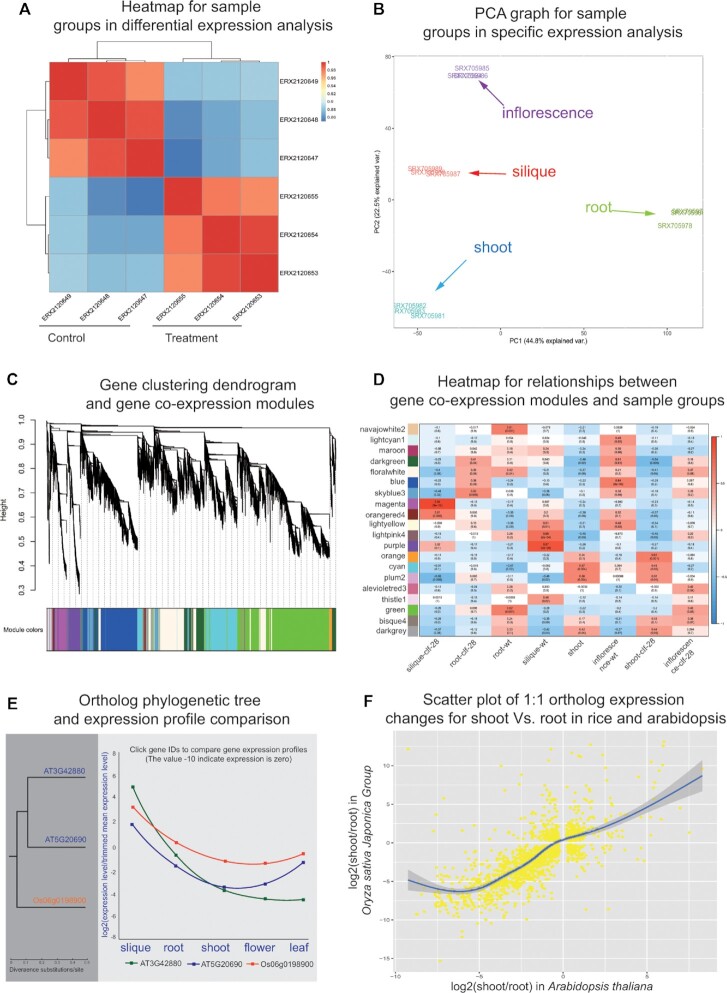
Figure 3.
Examples of result visualization generated by online analysis tools. (A) Sample clustering heatmap based on gene expression profiles in differential expression analysis. (B) PCA plot of overall gene expression for samples covering four tissues in specific expression analysis. (C) Co-expression network analysis tool generates a gene clustering dendrogram corresponding to co-expression modules. (D) Heatmap relationship between gene co-expression modules and sample groups. (E) Cross-species expression conservation analysis tool generates a phylogenetic tree of 3 orthologs from rice and arabidopsis, and provides an interactive box to compare their expression profiles covering five tissues. The gene expression levels are log2 transformed ratios of the gene expression in a tissue divided by trimmed mean expression level. (F) Scatter plot of 1:1 ortholog gene expression change ratios for shoot Vs. root in rice and arabidopsis. The gene expression fold changes are log2 transformed values.