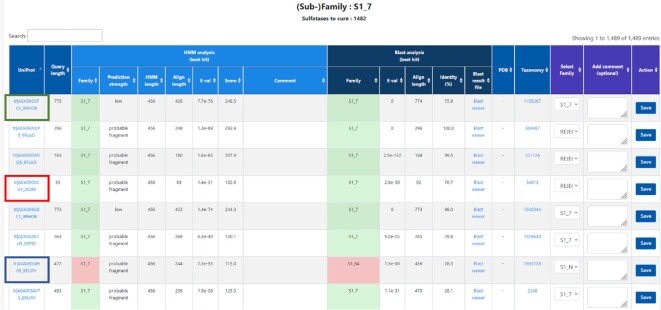
Figure 2.
Home page of the internal web-based curation tool. The updated pipeline automatically feeds the curation tool with candidate sulfatase sequences with uncertain status. Details of HMM and BLAST results are provided for each candidate sequence. The figure shows the results for the S1_7 subfamily. Three sequences are highlighted as example cases: (i) green box (UniProt: A0A086XT05_9RHOB): the HMM and BLAST analyses concur to an S1_7 assignation, but with HMM score slightly below 300 (248.5). Manual verification confirmed that this sequence contained a complete S1_7 sulfatase module but also hemolysin-type calcium-binding regions, explaining its larger size (775 residues). The S1_7 subfamily was thus confirmed in the ‘Select Family’ menu and saved with the ‘Action’ button. (ii) red box (A0A090XCV4_IXORI): the query is a very short sequence (93 residues) and is thus a sulfatase fragment (pseudo-gene or incorrectly predicted ORF) and is definitively rejected. (iii) blue box (A0A0B5GH05_9EURY): this sequence has the correct size to be a functional S1 sulfatase (472 residues) but the HMM and BLAST analyses do not concur on the same subfamily assignment (S1_7 and S1_64). Therefore, this sulfatase is currently orphan and may be a seed for a future new subfamily. For now, it is assigned to the S1_NC subfamily.