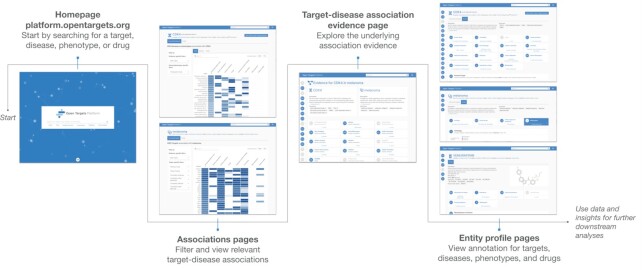
Figure 1.
A journey through the Platform web interface. The Open Targets Platform web interface is the first point of access for most users, and was completely redesigned to create the Next Generation Platform. The unified search box is connected to a series of tools allowing users to query different therapeutic hypotheses. From the homepage, users can navigate to association pages, with prioritised lists of target–disease associations. From there, users can access target–disease evidence pages, detailing the available evidence for an association. Once the evidence for a target–disease association has been assessed, users can explore entity profile pages, containing annotation information for each target, disease/phenotype and drug in the Platform to further build their hypothesis. For targets, this includes investigating whether it is expressed in a suitable tissue, what type of modality may be suitable and whether modulation is likely to be safe, whether there are already known drugs or available chemical probes for validation experiments, and whether interacting proteins may be more suitable targets. For disease/phenotype, the user can investigate known drugs and their targets, or explore targets associated with disease phenotypes through ontology expansion. Drug annotation pages provide a user with the mechanism of action and safety information related to modulating a target.