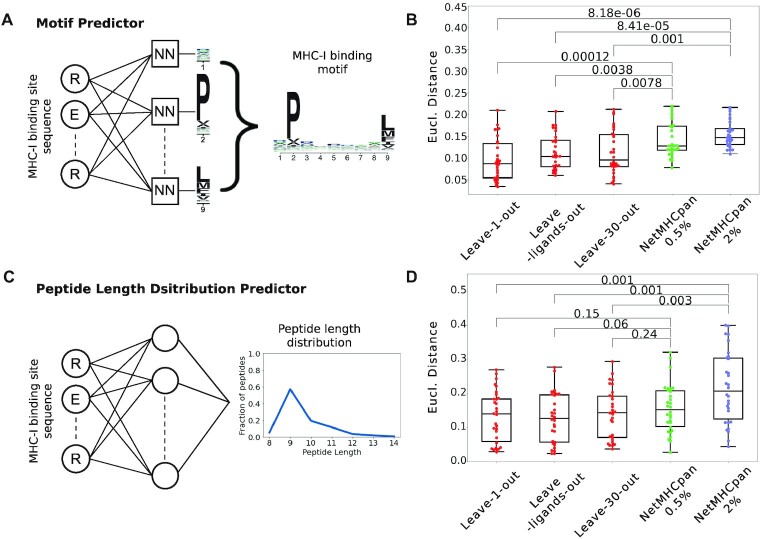
Figure 3.
Predicting the binding properties of MHC-I molecules. (A) Machine learning framework for the prediction of binding motifs for MHC-I alleles. Distinct neural networks (NN) were built for each peptide length and each position, and the final motif for a given peptide length is built by combining all position. The example illustrates the neural networks for predicting 9-mer binding motifs. (B) Leave-one-allele-out, leave-ligands-out and leave-30-alleles-out cross validation of the predictions of binding motifs for the 30 MHC-I alleles that are not part of the training set of NetMHCpan. Predicted motifs from our method and from NetMHCpan4.1 (at percentile ranks smaller than 0.5% or 2%) were compared to the experimental ones using the Euclidean distance. (C) Architecture of the neural network for the predictions of peptide length distributions for MHC-I alleles without experimental ligands. (D) Leave-one-allele-out, leave-ligands-out and leave-30-alleles-out cross validation of the predictions of peptide length distributions for the 30 MHC-I alleles that are not part of the training set of NetMHCpan. Predicted peptide length distributions from our method and from NetMHCpan4.1 (at percentile ranks smaller than 0.5% or 2%) were compared to the experimental ones using the Euclidean distance. Boxplots indicate the median, upper and lower quartiles. P-values were computed with the paired two-sided Mann–Whitney U-test.