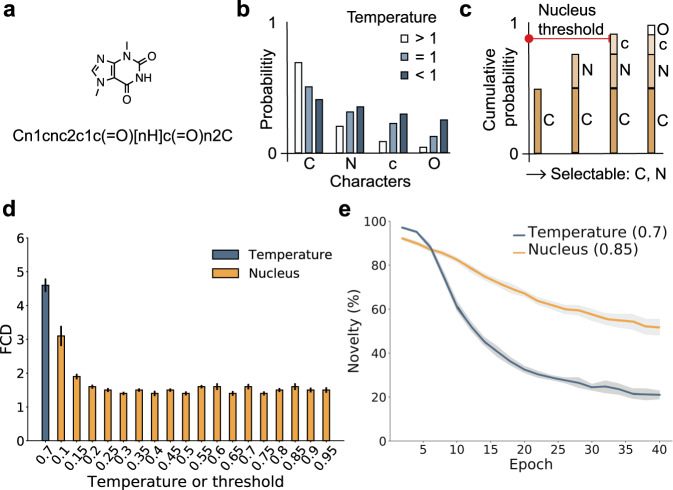
Fig. 1. De novo molecular generation with the CLM.
a SMILES string representation of a molecule. b Example of the effect of the temperature parameter on the probability distribution learnt by the CLM. c Example of the effect of the nucleus sampling threshold. Only the characters N and C can be sampled here. d Fréchet ChemNet Distance (FCD) comparison between temperature and nucleus sampling after the pretraining (reported as the mean with standard deviation over 10 repeats with 5000 molecules sampled per repeat). e Comparison of the novelty of the generated SMILES strings during the transfer learning between temperature sampling (temperature = 0.7) and nucleus sampling (threshold = 0.85). Mean values (lines) and standard deviations (shaded areas) are shown for ten repeats (1000 SMILES strings were sampled every second epoch over 40 epochs). Novelty is expressed as the percentage of SMILES strings generated that were valid and not included in either the training or the fine-tuning data.