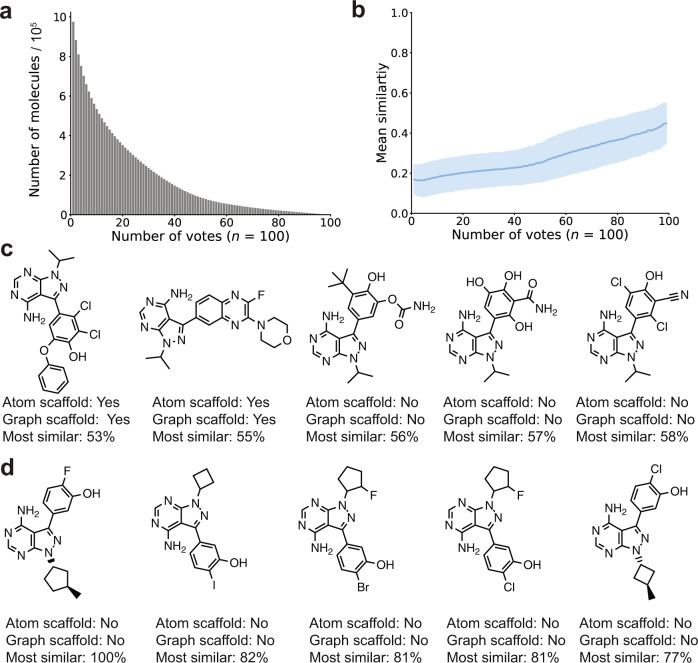
Fig. 3. Molecule ranking with a deep ensemble model.
a Number of molecules in the refined virtual chemical library that were predicted as “highly active” as a function of the number of votes (confidence level). b Average structural similarity (Tanimoto similarity index computed on Morgan fingerprints) of each de novo design to the fine-tuning set as a function of the number of votes. The solid line represents the mean value, with the shaded area representing the standard deviation. c Top-ranked designs (99/100 votes) selected with the most distant nearest neighbor, whose similarity is indicated below the structure (“Most similar”) in the fine-tuning set. The atom (“Atom scaffold”) and graph (“Graph scaffold”) scaffold novelty of the structure with respect to the fine-tuning set is indicated below each structure (“Yes”: new, “No”: not new). d Top-ranked designs (99/100 votes) selected with the closest nearest neighbor in the fine-tuning set.