Figure 2: Application of LotAN K6 specificity for UbiCRest analysis.
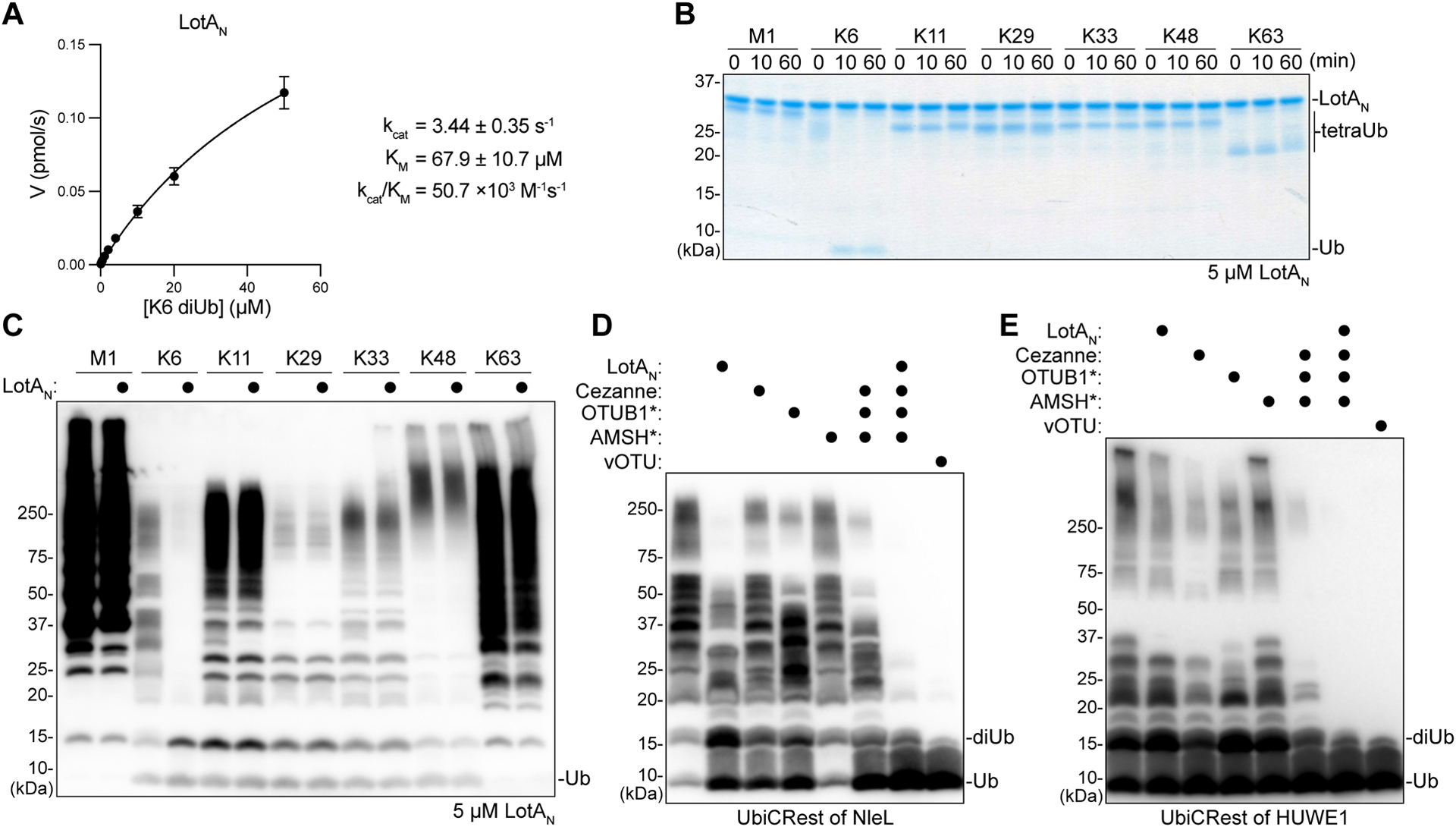
A. Kinetic parameters of LotAN (1–300) measured by changes in fluorescence polarization of labeled K6 diUb. Initial rates of diUb cleavage were measured over a range of substrate concentrations and fit to a Michaelis-Menten model. Error bars represent standard deviation over three measurements, each made in triplicate.
B. Gel-based specificity analysis against seven tetraUb linkages. Reactions containing a high concentration (5 μM) of LotAN were sampled at the indicated timepoints, quenched, and resolved by SDS-PAGE with Coomassie staining.
C. Homogeneous assemblies of seven polyUb linkage types were treated with a high concentration (5 μM) of LotAN for 2 h before the reactions were quenched and visualized by anti-Ub Western blot.
D. UbiCRest analysis of an NleL ligase assembly with 1 μM K6-specific LotAN, K11-specific Cezanne, K48-specific OTUB1*, K63-specific AMSH*, nonspecific vOTU, or the indicated combinations. Reactions were visualized by anti-Ub Western blot. Cleavage of NleL-assembled polyUb can be observed by a decrease in the “smear” or by a reappearance of monoUb.
E. UbiCRest analysis of a HUWE1 ligase assembly with 1 μM K6-specific LotAN, K11-specific Cezanne, K48-specific OTUB1*, K63-specific AMSH*, nonspecific vOTU, or the indicated combinations. Reactions were visualized by anti-Ub Western blot. Cleavage of HUWE1-assembled polyUb can be observed by a decrease in the “smear” or by a reappearance of monoUb.
See also Figure S2.
