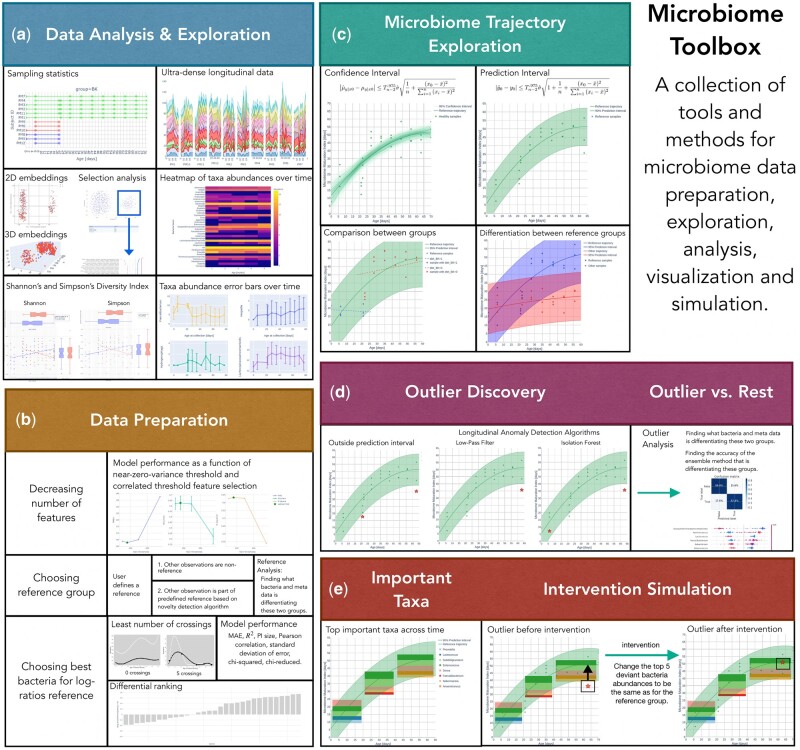
Fig. 1.
Microbiome Toolbox has multiple components for (a) exploration of microbiome data, (b) preparing data for subsequent analyses, (c) constructing microbiome trajectories, (d) determining who is an outlier or within a trajectory and (e) identifying important features as possibilities to return an outlier sample back on to the trajectory. The data used to illustrate the functionalities of this Microbiome Toolbox is from the R-package—metagenomeSeq (Paulson et al., 2013). Briefly, 12 germ-free mice fed a low-fat, plant polysaccharide-rich diet, were inoculated with adult human fecal material by gavage. Mice remained on the same diet for 4 weeks before a subset of six mice were switched to a high-fat, high-sugar diet for an additional 8 weeks