Fig. 1.
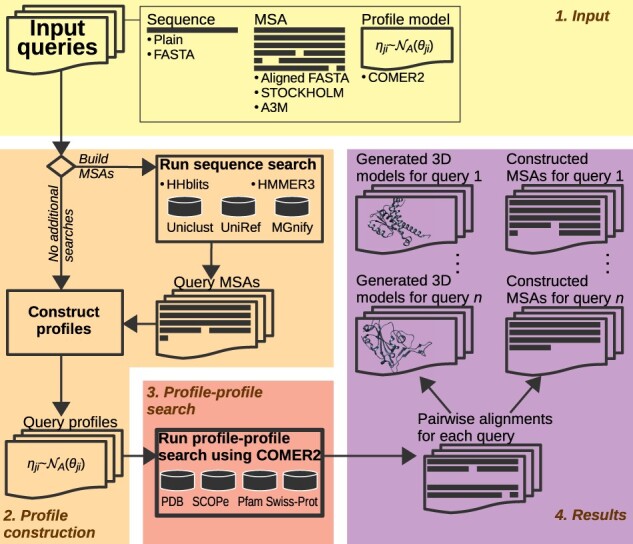
Flowchart for the COMER web server. (1) The user can provide sequences, MSAs and COMER2 profiles in different formats in the same input field. The server automatically determines the format. (2) The server can be instructed to build informative MSAs for user queries (profiles excluded). In this case, the server performs additional sequence searches using HHblits (Remmert et al., 2011; Steinegger et al., 2019a) against the Uniclust (Mirdita et al., 2017) or BFD (Steinegger et al., 2019b) database (not shown) and/or HMMER3 (Eddy, 2011) against the UniRef (Suzek et al., 2015) or MGnify (Mitchell et al., 2020) sequence database. COMER2 profiles are constructed for each sequence and MSA corresponding to a user query. (3) Searching at various levels of protein knowledge is provided by a profile–profile search across different COMER2 databases (several are shown; see text for details). (4) Based on the produced alignments, the user can construct profile–profile-guided MSAs and generate structural models by homology in bulk
