Fig. 3.
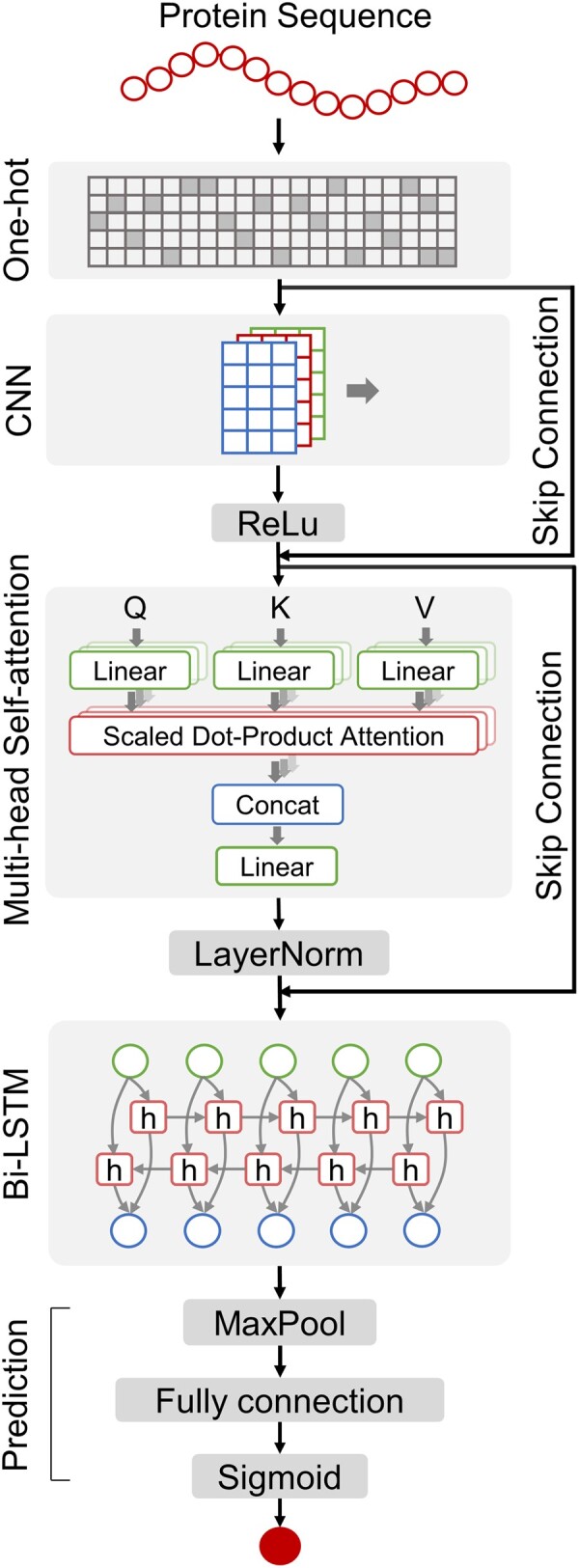
DeepCellEss framework. DeepCellEss accepts a protein sequence as input and converts it into a numerical matrix using one-hot encoding. After that, a CNN module is employed to effectively capture sequence local information. The multi-head self-attention is used to produce residue-level attention scores for model interpretability. Additionally, two skip-connection operations are implemented around CNN and the multi-head self-attention to avoid the model degradation problem. After multi-head self-attention, a bi-LSTM module is applied to model sequential data by learning long-range dependencies. Finally, the prediction task is performed after a max-pooling and fully connected layer
