Fig. 2.
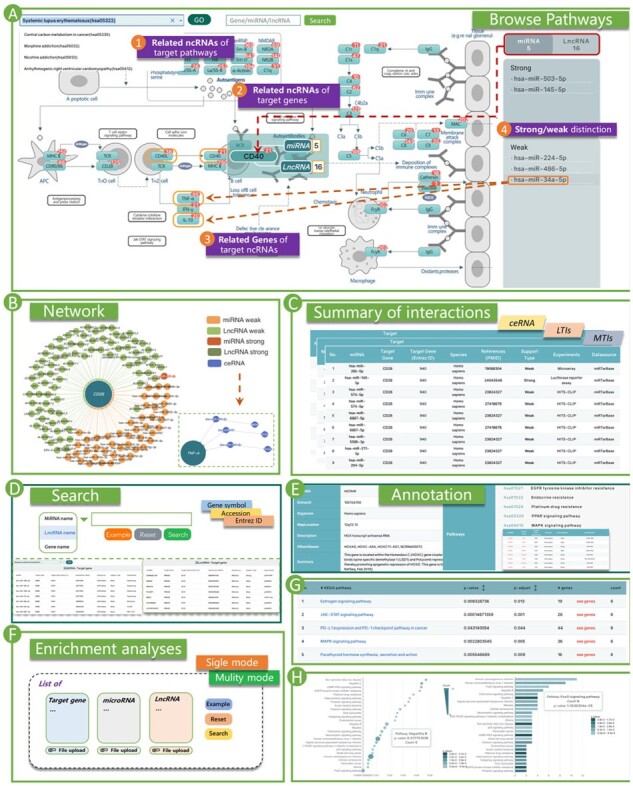
Introduction and usage of NcPath. (A) By browsing the pathway ‘Systemic lupus erythematosus’, users can intuitively obtain (i) related ncRNAs of target pathways; (ii) related ncRNAs of each target gene; (iii) related genes of target ncRNAs; and (iv) strong/weak distinction. (B) The non-coding-related interaction network of the gene CD28 and the ceRNA network for the gene TNF-A. (C) The table summarizing the interactions of the gene CD28 in the pathway ‘Systemic lupus erythematosus’, including MTIs, LTIs and ceRNA information. (D) Users can search by gene and non-coding RNA to obtain comprehensive information, and intuitively browse MTIs, LTIs and ceRNA networks related to the target pathway. (E) The display containing annotation information, associated pathways and non-coding RNAs–target interaction tables from the search function. (F) NcPath provides two input modes of enrichment analysis, including the single-class and multi-class modes. (G) Table with the results of the enrichment analysis. (H) Bubble results and column charts of enrichment analysis
