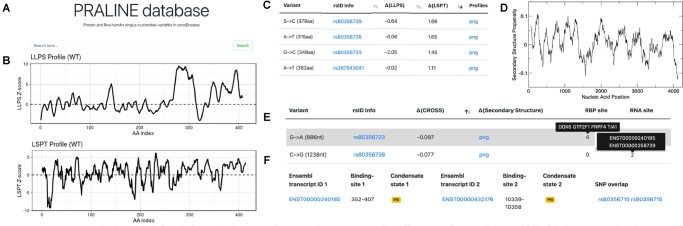
Fig. 1.
PRALINE database. (A) Search bar. The input can be a protein or an RNA in different ID formats. (B) Liquid–liquid phase separation (LLPS) and liquid–solid phase transition (LSPT) propensity profiles of a protein are predicted using catGRANULE and Zyggregator algorithms. (C) Protein SNVs description table: the difference in LLPS and LSPT compared to the WT is provided. (D) CROSS secondary structure propensity profile image of a RNA sequence. (E) RNA SNVs description table: the difference in CROSS secondary structure propensity compared to the WT, corresponding to a 11-nt window around the mutation, is provided, as well as proteins and RNAs interacting with the query transcript containing the SNV. (F) Example of an RNA–RNA interaction table. The information about RNAs’ binding sites, condensates localization and SNVs falling inside at least one of the binding sites are reported. The examples B–F relate to TARDBP