Figure 3.
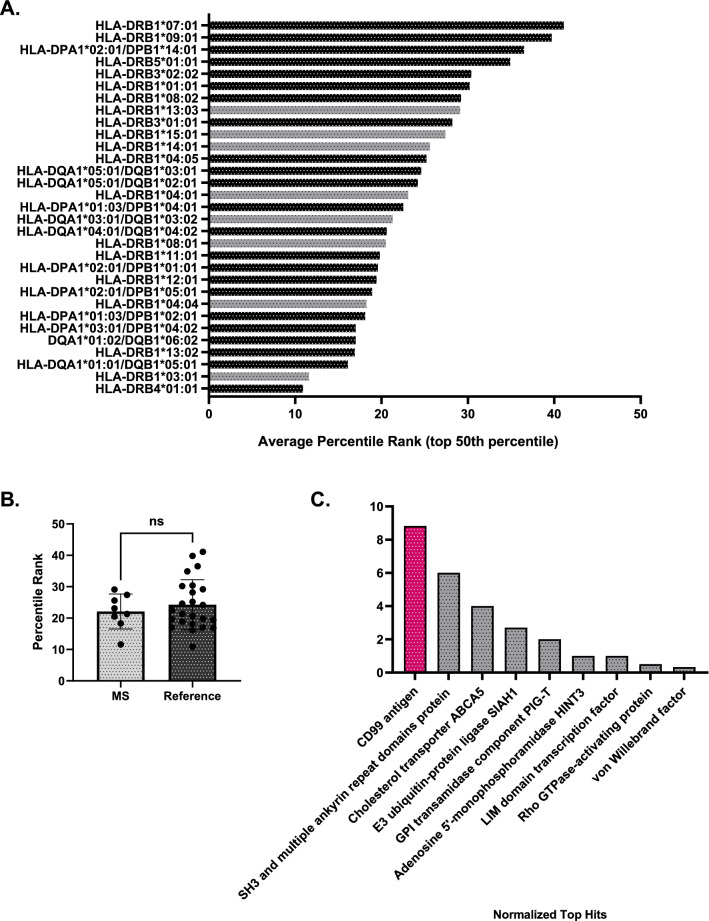
MHC-binding prediction provides insight on proteins which may engage in molecular mimicry beyond PLP. (A) MHC-binding prediction tool by the IEDB was used to determine the binding of nucleocapsid peptide hits from the PEPMatch analysis and MS-associated alleles (gray) versus a reference set of alleles (black)25. Shown are the top 50th percentile of average binding predictions of all 15mer nucleocapsid peptides from the original analysis and their respective HLA alleles that are predicted to bind. The lower the percentile rank, the stronger the predicted binding between the allele and the set of peptides run in the analysis. (B) Data from (A) grouped by allele classification. (C) Peptide:allele combinations with a percentile rank of 10th or less were collected and assessed for their respective MS-associated protein matches from the original PEPMatch analysis; plotted are the proteins that were returned from this analysis that are normalized to their respective number of peptides input.