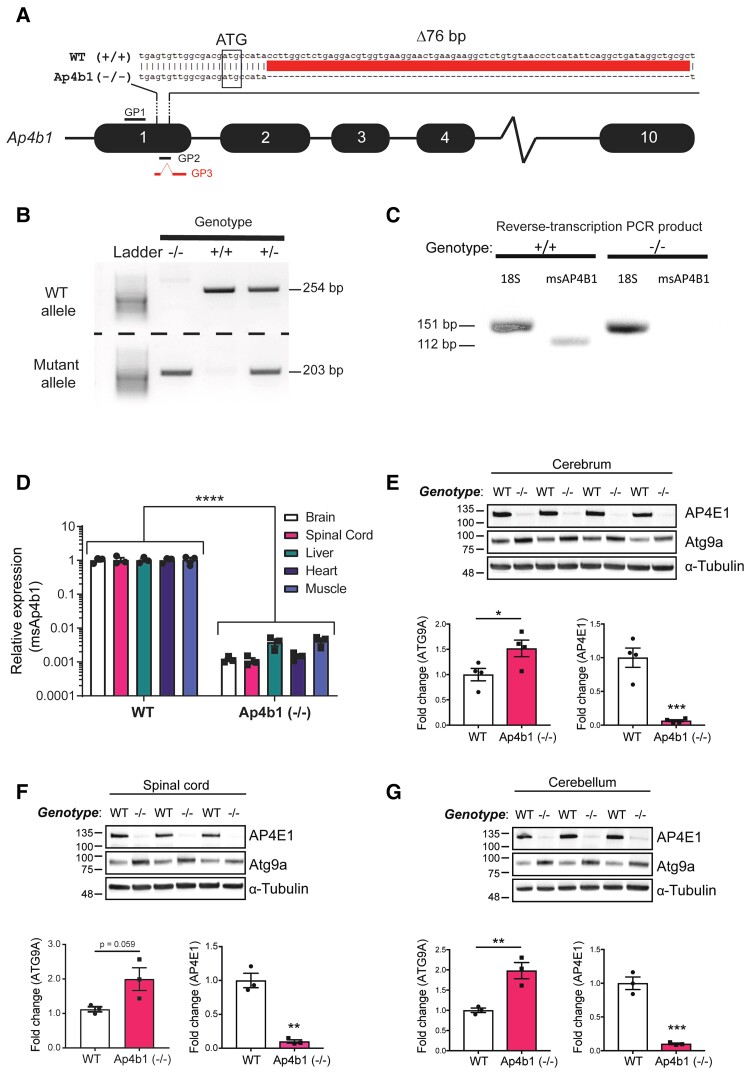
Figure 1.
Genetic characterization of the C57BL/6J-Ap4b1em5Lutzy/J mouse model of SPG47. Schematic of CRISPR-mediated murine Ap4b1 gene knockout. Guide strand directed DNA cleavage resulted in a 76 bp genomic DNA deletion downstream of the ATG start codon in Exon 1. This deletion is predicted to result in a frameshift leading to a premature stop codon and a non-functional truncated protein. Genotyping primer binding sites are shown as GP1 (forward primer), GP2 (reverse primer), and GP3 (reverse primer) (A). PCR-based genotyping of genomic DNA extracted from mouse tail tissue. Two separate PCR reactions generate products for either the WT (254 bp; product from GP1 and GP2 primers) or mutant (Ap4b1 (−/−)) (203 bp; product from GP1 and GP3 primers) allele (B). RT-PCR of total RNA extracted from WT (+/+) and Ap4b1 (−/−) mouse brain tissue using primers for murine Ap4b1 (msAP4b1). 18S primers were used as a control (C). RT-qPCR of total RNA extracted from both peripheral and CNS tissues showing expression of murine Ap4b1 in Ap4b1(−/−) mice compared to WT mice (D). Western blot detection of AP-4 ɛ (AP4E1) and Atg9a protein expression in Ap4b1(−/−) mouse tissue compared to WT, from cerebrum (E), spinal cord (F) and cerebellum (G). Data presented as mean ± S.E.M, analysed by Student’s two-tailed t-test (E–G) or two-way ANOVA with Bonferroni post hoc test for multiple comparisons (D). n ≥ 3 animals per group, all animals analysed in this figure were male. *P < 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001. See Supplementary material (Supplementary Figure 3) for uncropped gel of Fig. 1B. See Supplementary material (Supplementary Figure 4) for uncropped blots of Fig. 1E–G.