Fig. 5.
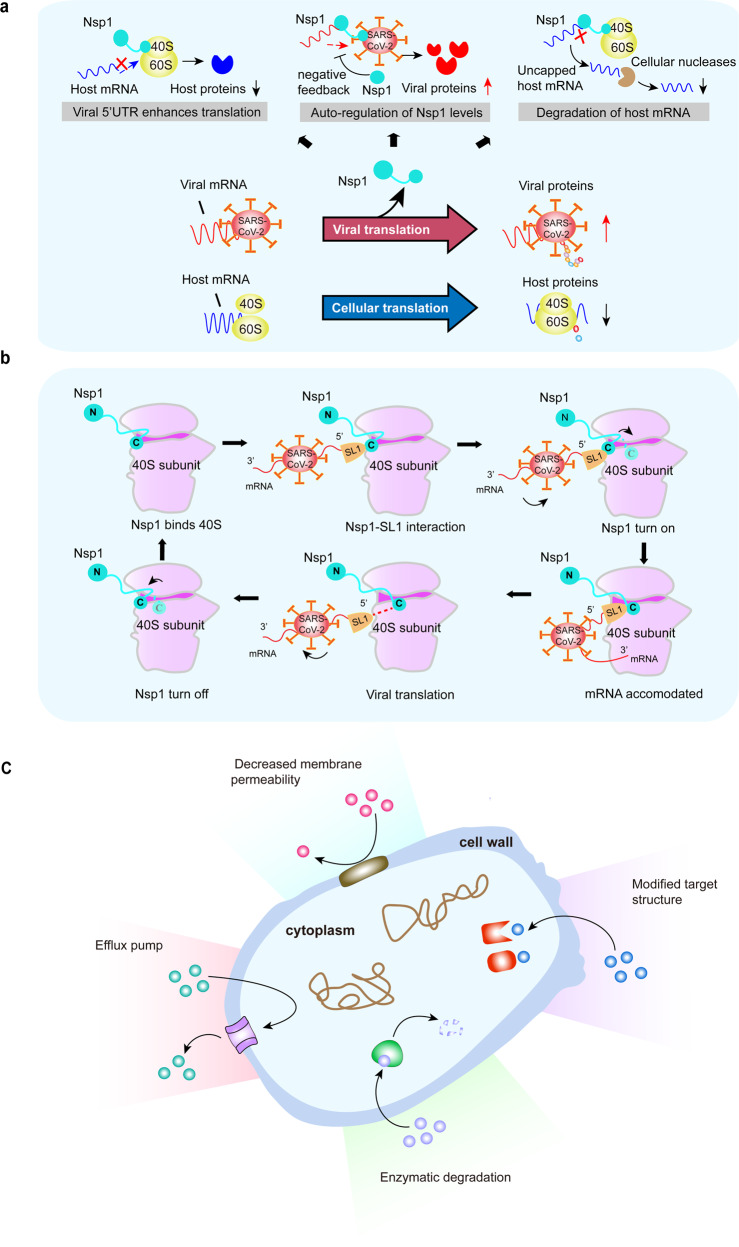
a SARS-CoV-2 regulates ribosome biogenesis in host cells through multiple pathways. Nsp1 is used by the SARS-CoV-2 virus to ensure its own replication and spread in the human host. The 5’UTR of viral Nsp1 is a key factor in directing ribosomes to viral transcripts and blocking host cell mRNA translation. Nsp1 blocks host mRNA translation and enhance the synthesis of viral proteins. Nsp1 alters the balance between viral and host cellular mRNAs through the cleavage of host mRNAs, leading to the degradation of host mRNAs by cellular nucleases. b Nsp1 acts as a gatekeeper to help SARS-CoV-2 evasion. Early in infection, Nsp1 binds to the 40S subunit with the carboxy-terminal domain of Nsp1. The viral mRNA transcript forms a translation initiation complex with the 40S-Nsp1 complex in its 5’UTR SL1. The carboxy-terminal domain of Nsp1 is removed to open the ribosome biogenesis channel for viral mRNA. The translation initiation, elongation, and termination further proceed. Upon termination, the viral mRNA is released, and the Nsp1 carboxy terminus refolds to prevent any de novo translation of cellular mRNA. c Mechanisms of bacterial resistance. Reducing the concentration of the harmful drug in the bacterium by decreasing the permeability of the bacterial outer membrane. The antimicrobial substance in the bacterial cell is removed by the efflux pumps. Mutation or modification of the target structure to reduce the affinity for antibiotics. Enzymatic degradation of antibiotics. Nsp1 nonstructural protein 1