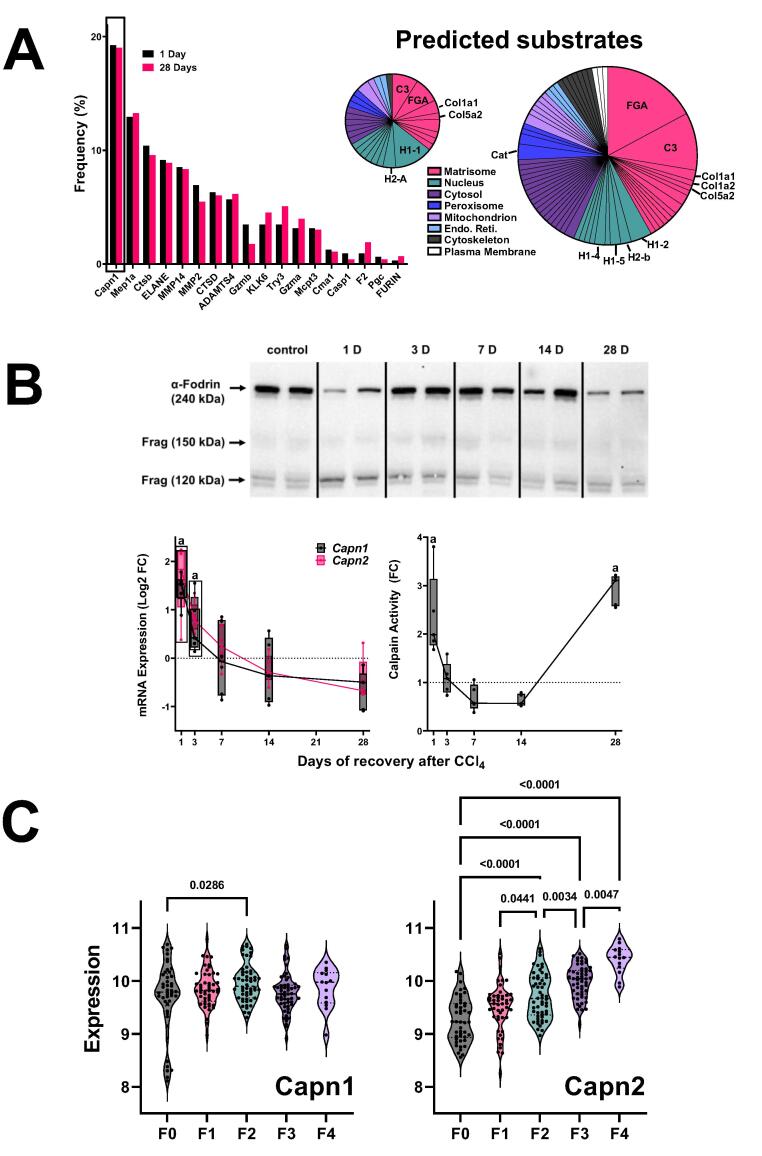
Fig. 6.
Expression and activity of predicted protease from ProteaSix. Panel A: Frequency distribution of predicted proteases 1 (black) and 28 (red) days after CCl4, as determined by ProteaSix (left). The type of parent proteins identified a potential substrates for Capn1/2 cleavage by ProteaSix, organized by subcellular localization (right). The relative size of the pie charts (1 D vs 28 D) is reflective of the amount of peptides identified at each timepoint. Panel B: Western blot analysis of α-fodrin (upper). Representative bands are shown from experiments repeated at least 4 times. The 240 kDa band represents intact α-fodrin, while the 150 and 120 kDa bands represent calpain-cleaved fragments. The expression (lower, left) of Capn1 and Capn1 were determined by real-time rtPCR. The activity of calpain in hepatic extracts (lower right) was determined by ELISA. Panel C: the expression of Capn1 and Capn2 in human NASH as a function of fibrosis score (METAVIR) are shown. Quantitative data are reported either as box and whisker plots (Panel B), which show the median (thick line), interquartile range (box) and range (whiskers) (n = 4–6). (n = 6–8; panel B), or as violin plots, which show the median (thick line), interquartile range (thin lines) and frequency distribution of the data (Panel C). aP < 0.05 compared with absence of CCl4 by 1-way ANOVA using Bonferroni’s post hoc test (panel B). P-values for human NASH fibrosis by ANOVA using Bonferroni’s post hoc test are shown between bracketed groups (panel C). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)